Gene
KWMTBOMO03090
Annotation
PREDICTED:_uncharacterized_protein_K02A2.6-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.228
Sequence
CDS
ATGTTCGGGACGTCTTCTTTAAGTGTTTTGTCGAGTTTTGATCATCAAAATCAAGATTGGAAGACATATAAGAGTAGATTGCAGCAATGGTGCATAGCTAACGGCATCGATGAGCAGTCTGATAAAGCCGGAATCAAGAGAAGAGCAGTACTCCTCAGCGCACTCACAGAGAGTACCTACCAGCTCGCAAGCAACTTAGCGTTACCCGACGATATTGCGACAAAGAGCTATGAAAGCATTTTAGGTATTTTAGATGAACACTTTATTCCAAAGCGGTGCGGATTCGCAGAGCGGTATAATTTTTACGCAGCGATGCAGCAAAATGGTGAAACTCATGGGCAATGGGCCGCACGCTTGCGAGGTTTGGCCGCACACTGCGCTTTCAAAAACCTAGAAGAATCTCTGCTAGATAAATTCGTCATGGGAATGTTGCGAGGACCCGAGCGAGAAAGGCTTTTCGCACAAGAAATCAGAGAGCTGACGTTGAGCAAAGCTGTGGAGCTGGCAGAGAGCGTGCGGTGCGCACGAGAAGGTGCAGCTGCAGCGGTAGTGCCGGAATCATCCGGTCGGGAAGGCGTTTTCAAGATTGCCCAACAAGCAAAAGGGAGCGCGGACCGAACAGCCGGCGTAAAATGCTTAGTATGTGGTTACAAAAATCACAAAACGTCAGAGTGCCGATTTTCTAAATTTAAGTGCAGAATATGTGATCGTAAAGGTCATCTCCGTAGAATGTGCCCCAGGAACAAAGTGAAGCACTTGGAGCAAGGTAACGACGATGACGAGGGCGATGACGGTGAGTTTTTGAAAAATATAAGTTGTCCAAATGGTGAGCCTTTGTACGAGACCGTGTGTTTAGAAGGTCGTAAATTTAGATGCGAAATTGACAGTGGTTCCCCTGTGTTCTGTTATATCCGAAAAGACATACAAGCGTTGGTTTTCCGGGGTTCCGTTGTCACCACCAGCGAAAAAATTATTTACGTATGA
Protein
MFGTSSLSVLSSFDHQNQDWKTYKSRLQQWCIANGIDEQSDKAGIKRRAVLLSALTESTYQLASNLALPDDIATKSYESILGILDEHFIPKRCGFAERYNFYAAMQQNGETHGQWAARLRGLAAHCAFKNLEESLLDKFVMGMLRGPERERLFAQEIRELTLSKAVELAESVRCAREGAAAAVVPESSGREGVFKIAQQAKGSADRTAGVKCLVCGYKNHKTSECRFSKFKCRICDRKGHLRRMCPRNKVKHLEQGNDDDEGDDGEFLKNISCPNGEPLYETVCLEGRKFRCEIDSGSPVFCYIRKDIQALVFRGSVVTTSEKIIYV
Summary
Similarity
Belongs to the complex I LYR family.
Uniprot
Q5KTM2
A0A2H1X402
A0A2A4JWI9
A0A2W1BSI8
A0A0L7KQ56
A0A0L7K2P5
+ More
A0A0L7K501 A0A0A9YE87 A0A146MBD7 A0A1Y1KIG3 A0A146M835 X1WRS2 X1WQM8 J9KQR5 A0A1Y1KCE7 A0A146L8C3 X1XHW3 A0A1Y1KPQ4 X1WXW3 J9LDN7 J9KFN3 J9KYF7 J9KL56 K7EWQ6 A0A2R7X643 J9JNK8 J9KZ86 X1XT86 A0A2R7X8N1 A2TGR4 A0A2B4RXD0 A0A2R7W917 A2TGR5 A0A2B4RTJ4 A0A3P8NG55 A0A3B3XP10 A0A0A9WT47 A0A3B1JY20 A0A3P8NHI5 A0A3P8QD94 A0A1Y1MAK2 A0A0V0UKI3 J9LBK5 A0A0V0UIV1 A0A2R7WLI7 W4LLX0 A0A0V0USX8 A0A0C2MSE0 A0A0V1BVI1 A0A0V0VH28 A0A1Y1K5W2 A0A1Y1LDW7 A0A2S2PLE8 A0A1Y1K4M7 A0A2B4R781 J9LID5 A0A0V1KNI7 A0A1Y1NDL2 A0A2S2N957 A0A1Y1NGB8 E5SDV0 L7MMK9 A0A146XMZ2 A0A3P8VSE7 A0A1B6DD07 A0A182GK90 A0A1B0CFC4 A0A0V0USW3 A0A1B6CPB9 A0A3P8NEQ7 A0A1A8C840 J9KM18 A0A0V1CV78 A0A1Y1NFA4 A0A1Y1LR51 A0A023G5Y2 X1WLT5 A0A0L7L8L8 A0A3B3H2N7 A0A1Y1N2Q0 A0A0V0VGS7 A0A1X7U2W2 A0A0V0VH72 A0A0V1NDZ1 A0A0V1N1F5 A0A1A8FCH6
A0A0L7K501 A0A0A9YE87 A0A146MBD7 A0A1Y1KIG3 A0A146M835 X1WRS2 X1WQM8 J9KQR5 A0A1Y1KCE7 A0A146L8C3 X1XHW3 A0A1Y1KPQ4 X1WXW3 J9LDN7 J9KFN3 J9KYF7 J9KL56 K7EWQ6 A0A2R7X643 J9JNK8 J9KZ86 X1XT86 A0A2R7X8N1 A2TGR4 A0A2B4RXD0 A0A2R7W917 A2TGR5 A0A2B4RTJ4 A0A3P8NG55 A0A3B3XP10 A0A0A9WT47 A0A3B1JY20 A0A3P8NHI5 A0A3P8QD94 A0A1Y1MAK2 A0A0V0UKI3 J9LBK5 A0A0V0UIV1 A0A2R7WLI7 W4LLX0 A0A0V0USX8 A0A0C2MSE0 A0A0V1BVI1 A0A0V0VH28 A0A1Y1K5W2 A0A1Y1LDW7 A0A2S2PLE8 A0A1Y1K4M7 A0A2B4R781 J9LID5 A0A0V1KNI7 A0A1Y1NDL2 A0A2S2N957 A0A1Y1NGB8 E5SDV0 L7MMK9 A0A146XMZ2 A0A3P8VSE7 A0A1B6DD07 A0A182GK90 A0A1B0CFC4 A0A0V0USW3 A0A1B6CPB9 A0A3P8NEQ7 A0A1A8C840 J9KM18 A0A0V1CV78 A0A1Y1NFA4 A0A1Y1LR51 A0A023G5Y2 X1WLT5 A0A0L7L8L8 A0A3B3H2N7 A0A1Y1N2Q0 A0A0V0VGS7 A0A1X7U2W2 A0A0V0VH72 A0A0V1NDZ1 A0A0V1N1F5 A0A1A8FCH6
Pubmed
EMBL
AB126055
BAD86655.1
ODYU01013266
SOQ59972.1
NWSH01000444
PCG76387.1
+ More
KZ149914 PZC78019.1 JTDY01007576 KOB65104.1 JTDY01013871 KOB52076.1 JTDY01011156 KOB57463.1 GBHO01013125 JAG30479.1 GDHC01002084 JAQ16545.1 GEZM01085308 JAV60010.1 GDHC01002706 JAQ15923.1 ABLF02030456 ABLF02003031 ABLF02009153 ABLF02009156 ABLF02036842 ABLF02042564 ABLF02052474 ABLF02006499 ABLF02041879 GEZM01092768 GEZM01092767 JAV56467.1 GDHC01014794 JAQ03835.1 ABLF02027007 ABLF02027010 ABLF02035806 GEZM01081409 JAV61645.1 ABLF02030002 ABLF02036937 ABLF02026287 ABLF02026295 ABLF02051114 ABLF02002867 ABLF02002868 ABLF02021819 ABLF02042177 ABLF02055259 ABLF02058160 AGCU01195578 KK857512 PTY27246.1 ABLF02001462 ABLF02041863 ABLF02066900 ABLF02066901 ABLF02026830 ABLF02012266 ABLF02012271 KK857520 PTY27255.1 EF199621 ABM90392.1 LSMT01000299 PFX20902.1 KK854477 PTY16206.1 EF199622 ABM90393.1 LSMT01000298 PFX20931.1 GBHO01031997 JAG11607.1 GEZM01036245 JAV82862.1 JYDK01000935 KRX51560.1 ABLF02038551 ABLF02038554 ABLF02042419 JYDK01000986 KRX51372.1 KK855004 PTY20231.1 AZHX01001899 ETW98899.1 JYDN01000181 KRX54109.1 JWZT01003307 KII67125.1 JYDH01000010 KRY40903.1 JYDN01000036 KRX62850.1 GEZM01096178 JAV55085.1 GEZM01061347 JAV70540.1 GGMR01017626 MBY30245.1 GEZM01096180 JAV55080.1 LSMT01001234 PFX12679.1 ABLF02030730 ABLF02030738 JYDW01000348 KRZ48953.1 GEZM01005739 JAV95951.1 GGMR01001051 MBY13670.1 GEZM01005740 JAV95950.1 GACK01000385 JAA64649.1 GCES01042893 JAR43430.1 GEDC01013717 JAS23581.1 JXUM01013128 KQ560369 KXJ82759.1 AJWK01009946 KRX54093.1 GEDC01021971 JAS15327.1 HADZ01011823 HAEA01016148 SBP75764.1 ABLF02038615 ABLF02064938 JYDI01000102 KRY52595.1 GEZM01009753 JAV94337.1 GEZM01050895 JAV75318.1 GBBM01006246 JAC29172.1 ABLF02018110 ABLF02065955 JTDY01002326 KOB71659.1 GEZM01014356 JAV92134.1 KRX62729.1 KRX62859.1 JYDM01000372 KRZ82101.1 JYDO01000016 KRZ77842.1 HAEB01010230 SBQ56757.1
KZ149914 PZC78019.1 JTDY01007576 KOB65104.1 JTDY01013871 KOB52076.1 JTDY01011156 KOB57463.1 GBHO01013125 JAG30479.1 GDHC01002084 JAQ16545.1 GEZM01085308 JAV60010.1 GDHC01002706 JAQ15923.1 ABLF02030456 ABLF02003031 ABLF02009153 ABLF02009156 ABLF02036842 ABLF02042564 ABLF02052474 ABLF02006499 ABLF02041879 GEZM01092768 GEZM01092767 JAV56467.1 GDHC01014794 JAQ03835.1 ABLF02027007 ABLF02027010 ABLF02035806 GEZM01081409 JAV61645.1 ABLF02030002 ABLF02036937 ABLF02026287 ABLF02026295 ABLF02051114 ABLF02002867 ABLF02002868 ABLF02021819 ABLF02042177 ABLF02055259 ABLF02058160 AGCU01195578 KK857512 PTY27246.1 ABLF02001462 ABLF02041863 ABLF02066900 ABLF02066901 ABLF02026830 ABLF02012266 ABLF02012271 KK857520 PTY27255.1 EF199621 ABM90392.1 LSMT01000299 PFX20902.1 KK854477 PTY16206.1 EF199622 ABM90393.1 LSMT01000298 PFX20931.1 GBHO01031997 JAG11607.1 GEZM01036245 JAV82862.1 JYDK01000935 KRX51560.1 ABLF02038551 ABLF02038554 ABLF02042419 JYDK01000986 KRX51372.1 KK855004 PTY20231.1 AZHX01001899 ETW98899.1 JYDN01000181 KRX54109.1 JWZT01003307 KII67125.1 JYDH01000010 KRY40903.1 JYDN01000036 KRX62850.1 GEZM01096178 JAV55085.1 GEZM01061347 JAV70540.1 GGMR01017626 MBY30245.1 GEZM01096180 JAV55080.1 LSMT01001234 PFX12679.1 ABLF02030730 ABLF02030738 JYDW01000348 KRZ48953.1 GEZM01005739 JAV95951.1 GGMR01001051 MBY13670.1 GEZM01005740 JAV95950.1 GACK01000385 JAA64649.1 GCES01042893 JAR43430.1 GEDC01013717 JAS23581.1 JXUM01013128 KQ560369 KXJ82759.1 AJWK01009946 KRX54093.1 GEDC01021971 JAS15327.1 HADZ01011823 HAEA01016148 SBP75764.1 ABLF02038615 ABLF02064938 JYDI01000102 KRY52595.1 GEZM01009753 JAV94337.1 GEZM01050895 JAV75318.1 GBBM01006246 JAC29172.1 ABLF02018110 ABLF02065955 JTDY01002326 KOB71659.1 GEZM01014356 JAV92134.1 KRX62729.1 KRX62859.1 JYDM01000372 KRZ82101.1 JYDO01000016 KRZ77842.1 HAEB01010230 SBQ56757.1
Proteomes
PRIDE
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR034128 K02A2.6-like
IPR000477 RT_dom
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR032710 NTF2-like_dom_sf
IPR018222 Nuclear_transport_factor_2_euk
IPR041373 RT_RNaseH
IPR008777 Phytoreo_Pns
IPR008011 Complex1_LYR
IPR005162 Retrotrans_gag_dom
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR034128 K02A2.6-like
IPR000477 RT_dom
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR032710 NTF2-like_dom_sf
IPR018222 Nuclear_transport_factor_2_euk
IPR041373 RT_RNaseH
IPR008777 Phytoreo_Pns
IPR008011 Complex1_LYR
IPR005162 Retrotrans_gag_dom
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
Gene 3D
ProteinModelPortal
Q5KTM2
A0A2H1X402
A0A2A4JWI9
A0A2W1BSI8
A0A0L7KQ56
A0A0L7K2P5
+ More
A0A0L7K501 A0A0A9YE87 A0A146MBD7 A0A1Y1KIG3 A0A146M835 X1WRS2 X1WQM8 J9KQR5 A0A1Y1KCE7 A0A146L8C3 X1XHW3 A0A1Y1KPQ4 X1WXW3 J9LDN7 J9KFN3 J9KYF7 J9KL56 K7EWQ6 A0A2R7X643 J9JNK8 J9KZ86 X1XT86 A0A2R7X8N1 A2TGR4 A0A2B4RXD0 A0A2R7W917 A2TGR5 A0A2B4RTJ4 A0A3P8NG55 A0A3B3XP10 A0A0A9WT47 A0A3B1JY20 A0A3P8NHI5 A0A3P8QD94 A0A1Y1MAK2 A0A0V0UKI3 J9LBK5 A0A0V0UIV1 A0A2R7WLI7 W4LLX0 A0A0V0USX8 A0A0C2MSE0 A0A0V1BVI1 A0A0V0VH28 A0A1Y1K5W2 A0A1Y1LDW7 A0A2S2PLE8 A0A1Y1K4M7 A0A2B4R781 J9LID5 A0A0V1KNI7 A0A1Y1NDL2 A0A2S2N957 A0A1Y1NGB8 E5SDV0 L7MMK9 A0A146XMZ2 A0A3P8VSE7 A0A1B6DD07 A0A182GK90 A0A1B0CFC4 A0A0V0USW3 A0A1B6CPB9 A0A3P8NEQ7 A0A1A8C840 J9KM18 A0A0V1CV78 A0A1Y1NFA4 A0A1Y1LR51 A0A023G5Y2 X1WLT5 A0A0L7L8L8 A0A3B3H2N7 A0A1Y1N2Q0 A0A0V0VGS7 A0A1X7U2W2 A0A0V0VH72 A0A0V1NDZ1 A0A0V1N1F5 A0A1A8FCH6
A0A0L7K501 A0A0A9YE87 A0A146MBD7 A0A1Y1KIG3 A0A146M835 X1WRS2 X1WQM8 J9KQR5 A0A1Y1KCE7 A0A146L8C3 X1XHW3 A0A1Y1KPQ4 X1WXW3 J9LDN7 J9KFN3 J9KYF7 J9KL56 K7EWQ6 A0A2R7X643 J9JNK8 J9KZ86 X1XT86 A0A2R7X8N1 A2TGR4 A0A2B4RXD0 A0A2R7W917 A2TGR5 A0A2B4RTJ4 A0A3P8NG55 A0A3B3XP10 A0A0A9WT47 A0A3B1JY20 A0A3P8NHI5 A0A3P8QD94 A0A1Y1MAK2 A0A0V0UKI3 J9LBK5 A0A0V0UIV1 A0A2R7WLI7 W4LLX0 A0A0V0USX8 A0A0C2MSE0 A0A0V1BVI1 A0A0V0VH28 A0A1Y1K5W2 A0A1Y1LDW7 A0A2S2PLE8 A0A1Y1K4M7 A0A2B4R781 J9LID5 A0A0V1KNI7 A0A1Y1NDL2 A0A2S2N957 A0A1Y1NGB8 E5SDV0 L7MMK9 A0A146XMZ2 A0A3P8VSE7 A0A1B6DD07 A0A182GK90 A0A1B0CFC4 A0A0V0USW3 A0A1B6CPB9 A0A3P8NEQ7 A0A1A8C840 J9KM18 A0A0V1CV78 A0A1Y1NFA4 A0A1Y1LR51 A0A023G5Y2 X1WLT5 A0A0L7L8L8 A0A3B3H2N7 A0A1Y1N2Q0 A0A0V0VGS7 A0A1X7U2W2 A0A0V0VH72 A0A0V1NDZ1 A0A0V1N1F5 A0A1A8FCH6
Ontologies
GO
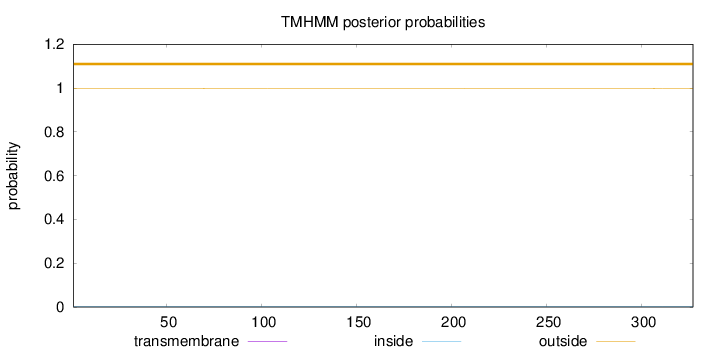
Topology
Length:
327
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0006
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.00291
outside
1 - 327
Population Genetic Test Statistics
Pi
6.780998
Theta
18.555303
Tajima's D
-1.381594
CLR
26.633334
CSRT
0.0734963251837408
Interpretation
Uncertain
