Gene
KWMTBOMO03087
Pre Gene Modal
BGIBMGA005815
Annotation
PREDICTED:_peptidyl-alpha-hydroxyglycine_alpha-amidating_lyase_1_[Bombyx_mori]
Full name
Peptidyl-alpha-hydroxyglycine alpha-amidating lyase 1
Alternative Name
Peptidylamidoglycolate lyase 1
dPAL1
dPAL1
Location in the cell
PlasmaMembrane Reliability : 1.881
Sequence
CDS
ATGAATAATTTATTATGCTTATTCAAAGTGATCGTTATGATACTGTGCTTAAGAGAATCAATTTACGTAAATGGATTTGGAGAGCGGGCCTATTTTTCAGACCGAGAAGACAATAATCCTGCATATTATATATCACCTGCATTGCAAAAACTAGAGCATGCACCAGAATTTGTGAAAGACTGGCCGAATATATCAATAACACTAGGCCAAGTTTCCGGAGTAACATTAGACAATGCAGGCAGGGTGCTAGTCTTCCACCGAGGTGAACATACTTGGGATGAGCACACATTTACAAATGGAAATATATACTTGGGAATTGGAAAACCAGCCATACAGCACAAAACTATTTTAATCTTCAATGAAACAGGAGAACTTCTTGATATGTGGGGTGATGATTTTTTTTATATGCCCCATGGCATAACGGTGGACTCTGAAGGTAATGTTTGGACCACTGATGTGGCACTACATCAAGTGTTCAAATTTAATGCTACCAACCGACGAGTGCCTGCTATGGTGCTGGGAGAAAAGTTTGTACCGGGAACAGATGACCATCATTTTTGTAAGCCAAGCGCGGTGGCCGTGTTGTCGACCGGTGACTTTTTCATAGCAGACGGCTACTGCAACCACAGGATCCTGAAATATAACCCTAAAGGGCAAATCATACTGCAGTGGGGTACTCATAACTCACACTCGCCGTTCTCTCTGAACGTGCCCCATGCGCTGGCGCTTGCTGAGAACTCGGGGGTGGTGCTGGTGGCGGACCGGGAGAGGGGCAGGGTCGCCAGCTTCAGGCACGACAACGGCTCCTACGTCACCTCCTACAGCAGCTGGCTCATGGGACCCAGGATCTTCAGTGTGGCCTATTCTCCGGTTCACGGAGGTCGTCTGTACGTGGTGAACGGGCCGAACGGCGTGGTCCCGGTGCGCGGCTACGTGCTGGAGCTGGCGTCCGGGCGGCTCGTGGGCACGTTCGACGCGCCCGCCGGCCTCGCCAGCCCGCACGACGTGGCGGCCGCGACCGACGGCTCCGCGGTCTACGTCGCGCAGCTCGCCCCGCACGCCGTCTACAAGTTCGTCGACGAGCGCCTGCGCGACGAGGCGCCGCCCGCACCCCCCGCACCCCCCGCGCCGCCCGCCGCGCCCCCCGCCACGCCCGCCACTCTCGGTGAGTAG
Protein
MNNLLCLFKVIVMILCLRESIYVNGFGERAYFSDREDNNPAYYISPALQKLEHAPEFVKDWPNISITLGQVSGVTLDNAGRVLVFHRGEHTWDEHTFTNGNIYLGIGKPAIQHKTILIFNETGELLDMWGDDFFYMPHGITVDSEGNVWTTDVALHQVFKFNATNRRVPAMVLGEKFVPGTDDHHFCKPSAVAVLSTGDFFIADGYCNHRILKYNPKGQIILQWGTHNSHSPFSLNVPHALALAENSGVVLVADRERGRVASFRHDNGSYVTSYSSWLMGPRIFSVAYSPVHGGRLYVVNGPNGVVPVRGYVLELASGRLVGTFDAPAGLASPHDVAAATDGSAVYVAQLAPHAVYKFVDERLRDEAPPAPPAPPAPPAAPPATPATLGE
Summary
Description
Probable lyase that catalyzes an essential reaction in C-terminal alpha-amidation of peptides. Mediates the dismutation of the unstable peptidyl(2-hydroxyglycine) intermediate to glyoxylate and the corresponding desglycine peptide amide. C-terminal amidation of peptides such as neuropeptides is essential for full biological activity.
Catalytic Activity
a [peptide]-C-terminal (2S)-2-hydroxyglycine = a [peptide]-C-terminal amide + glyoxylate
Cofactor
Zn(2+)
Biophysicochemical Properties
45 uM for peptidyl-alpha-hydroxyglycine
Similarity
Belongs to the peptidyl-alpha-hydroxyglycine alpha-amidating lyase family.
Keywords
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Lyase
Membrane
Metal-binding
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Zinc
Feature
chain Peptidyl-alpha-hydroxyglycine alpha-amidating lyase 1
Uniprot
H9J8H3
A0A2A4J8Y4
A0A2H1WA59
A0A0N0PBR4
A0A212EKT6
A0A194PQV8
+ More
A0A2J7QPM8 A0A2J7QPM1 A0A1D2NEY4 A0A067RHL7 A0A1Q3FGN2 A0A1Q3FGH4 A0A182KRQ4 A0A1A9ZCM1 B0VZE5 A0A182WIP2 A0A1A9X3N3 T1P8C3 A0A182M7E9 T1PM65 A0A1B0BH65 A0A182TDV8 A0A1A9YAZ3 A0A182RHT7 A0A1I8M5C6 A0A182P3M3 A0A182UW58 A0A1B0GDJ1 A0A084VWY7 Q7QJP6 A0A182IGW0 A0A1A9VP13 A0A182XZ69 A0A0L0BPQ8 A0A226F6T8 A0A1B6LWW9 A0A1B6G878 A0A182X3E7 C3YQ66 A0A1S3I9G3 A0A1S3IAE2 A0A1B6IDE5 A0A182GNQ6 A0A023ESW6 B4MRI5 A0A1I8PLY6 A0A182H9U8 B4J9H1 A0A182QCA2 A0A034W4J0 A0A182NHJ0 A0A0K8TYP3 A0A1W4UZC7 A0A0K8V2X3 Q17KT9 A0A168L575 A0A0A1XSQ6 A0A0K8WH17 W5JRY6 A0A0K8VGX3 A0A1W4UZ23 A0A2M3ZE61 A0A1W4V0N2 A0A2M3Z6W7 A0A0A1WQ78 A0A1Q1NAE3 A0A2M4BLU6 K7IX33 W8BY31 A0A0N8P0F3 B3N6T2 A0A0Q5WLM8 A0A0K2VHL2 A0A0B4LF14 B3MF37 B4LJX2 A0A0P8ZRB0 A0A0Q9WCX2 Q8I1H1 A0A2M4ANV6 B6IDN2 Q9V5E1 A0A224XRL7 A0A0M4EWG8 A0A182FI38 A0A0Q9X7Q6 G8EWC9 B4HMA5 A0A0R1DU55 A0A0J9RA86 B4KNT5 A0A0R1DMP8 B4NX21 T1KEL8 A0A154NX67 A0A0J9TXQ2 A0A3L8E0K9 A0A0K2VHQ9
A0A2J7QPM8 A0A2J7QPM1 A0A1D2NEY4 A0A067RHL7 A0A1Q3FGN2 A0A1Q3FGH4 A0A182KRQ4 A0A1A9ZCM1 B0VZE5 A0A182WIP2 A0A1A9X3N3 T1P8C3 A0A182M7E9 T1PM65 A0A1B0BH65 A0A182TDV8 A0A1A9YAZ3 A0A182RHT7 A0A1I8M5C6 A0A182P3M3 A0A182UW58 A0A1B0GDJ1 A0A084VWY7 Q7QJP6 A0A182IGW0 A0A1A9VP13 A0A182XZ69 A0A0L0BPQ8 A0A226F6T8 A0A1B6LWW9 A0A1B6G878 A0A182X3E7 C3YQ66 A0A1S3I9G3 A0A1S3IAE2 A0A1B6IDE5 A0A182GNQ6 A0A023ESW6 B4MRI5 A0A1I8PLY6 A0A182H9U8 B4J9H1 A0A182QCA2 A0A034W4J0 A0A182NHJ0 A0A0K8TYP3 A0A1W4UZC7 A0A0K8V2X3 Q17KT9 A0A168L575 A0A0A1XSQ6 A0A0K8WH17 W5JRY6 A0A0K8VGX3 A0A1W4UZ23 A0A2M3ZE61 A0A1W4V0N2 A0A2M3Z6W7 A0A0A1WQ78 A0A1Q1NAE3 A0A2M4BLU6 K7IX33 W8BY31 A0A0N8P0F3 B3N6T2 A0A0Q5WLM8 A0A0K2VHL2 A0A0B4LF14 B3MF37 B4LJX2 A0A0P8ZRB0 A0A0Q9WCX2 Q8I1H1 A0A2M4ANV6 B6IDN2 Q9V5E1 A0A224XRL7 A0A0M4EWG8 A0A182FI38 A0A0Q9X7Q6 G8EWC9 B4HMA5 A0A0R1DU55 A0A0J9RA86 B4KNT5 A0A0R1DMP8 B4NX21 T1KEL8 A0A154NX67 A0A0J9TXQ2 A0A3L8E0K9 A0A0K2VHQ9
EC Number
4.3.2.5
Pubmed
19121390
26354079
22118469
27289101
24845553
20966253
+ More
25315136 24438588 12364791 14747013 17210077 25244985 26108605 18563158 26483478 24945155 17994087 25348373 17510324 25830018 20920257 23761445 20075255 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 15198673 17550304 22936249 30249741
25315136 24438588 12364791 14747013 17210077 25244985 26108605 18563158 26483478 24945155 17994087 25348373 17510324 25830018 20920257 23761445 20075255 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 15198673 17550304 22936249 30249741
EMBL
BABH01027407
BABH01027408
BABH01027409
BABH01027410
BABH01027411
BABH01027412
+ More
NWSH01002324 PCG68525.1 ODYU01007288 SOQ49933.1 KQ460847 KPJ11991.1 AGBW02014213 OWR42099.1 KQ459597 KPI95129.1 NEVH01012088 PNF30531.1 PNF30532.1 LJIJ01000063 ODN03814.1 KK852678 KDR18657.1 GFDL01008332 JAV26713.1 GFDL01008400 JAV26645.1 DS231814 EDS31682.1 KA644956 AFP59585.1 AXCM01005430 KA649162 KA649170 AFP63799.1 JXJN01014195 CCAG010011915 ATLV01017808 KE525192 KFB42481.1 AAAB01008807 EAA04373.4 APCN01000215 JRES01001567 KNC21953.1 LNIX01000001 OXA65207.1 GEBQ01031647 GEBQ01023221 GEBQ01018526 GEBQ01011799 GEBQ01008105 GEBQ01003199 JAT08330.1 JAT16756.1 JAT21451.1 JAT28178.1 JAT31872.1 JAT36778.1 GECZ01026414 GECZ01011107 JAS43355.1 JAS58662.1 GG666540 EEN57570.1 GECU01022767 GECU01020157 GECU01017105 GECU01002011 JAS84939.1 JAS87549.1 JAS90601.1 JAT05696.1 JXUM01014692 KQ560410 KXJ82556.1 GAPW01001203 JAC12395.1 CH963850 EDW74724.2 JXUM01004851 JXUM01004852 KQ560182 KXJ83967.1 CH916367 EDW01452.1 AXCN02000593 GAKP01008431 JAC50521.1 GDHF01032923 JAI19391.1 GDHF01019067 JAI33247.1 CH477221 EAT47318.1 KU881664 ANC48002.1 GBXI01000372 JAD13920.1 GDHF01001970 JAI50344.1 ADMH02000632 ETN65499.1 GDHF01014150 JAI38164.1 GGFM01006038 MBW26789.1 GGFM01003500 MBW24251.1 GBXI01013719 JAD00573.1 KY011960 AQM52455.1 GGFJ01004884 MBW54025.1 AAZX01006819 GAMC01012391 JAB94164.1 CH902619 KPU76964.1 CH954177 EDV58181.2 KQS70322.1 HACA01032602 CDW49963.1 AE013599 AHN56057.1 EDV37665.2 CH940648 EDW61626.1 KPU76963.1 KRF80119.1 AY190939 AAO00999.1 GGFK01009140 MBW42461.1 BT050472 ACJ13179.1 ACL83084.2 AY051842 GFTR01005775 JAW10651.1 CP012524 ALC42145.1 CH933808 KRG04384.1 JF967750 AEI84584.1 CH480816 EDW47182.1 CM000157 KRJ98547.1 CM002911 KMY92624.1 EDW08980.1 KRJ98548.1 EDW89582.1 CAEY01000026 KQ434777 KZC04171.1 KMY92625.1 QOIP01000002 RLU25558.1 HACA01032603 CDW49964.1
NWSH01002324 PCG68525.1 ODYU01007288 SOQ49933.1 KQ460847 KPJ11991.1 AGBW02014213 OWR42099.1 KQ459597 KPI95129.1 NEVH01012088 PNF30531.1 PNF30532.1 LJIJ01000063 ODN03814.1 KK852678 KDR18657.1 GFDL01008332 JAV26713.1 GFDL01008400 JAV26645.1 DS231814 EDS31682.1 KA644956 AFP59585.1 AXCM01005430 KA649162 KA649170 AFP63799.1 JXJN01014195 CCAG010011915 ATLV01017808 KE525192 KFB42481.1 AAAB01008807 EAA04373.4 APCN01000215 JRES01001567 KNC21953.1 LNIX01000001 OXA65207.1 GEBQ01031647 GEBQ01023221 GEBQ01018526 GEBQ01011799 GEBQ01008105 GEBQ01003199 JAT08330.1 JAT16756.1 JAT21451.1 JAT28178.1 JAT31872.1 JAT36778.1 GECZ01026414 GECZ01011107 JAS43355.1 JAS58662.1 GG666540 EEN57570.1 GECU01022767 GECU01020157 GECU01017105 GECU01002011 JAS84939.1 JAS87549.1 JAS90601.1 JAT05696.1 JXUM01014692 KQ560410 KXJ82556.1 GAPW01001203 JAC12395.1 CH963850 EDW74724.2 JXUM01004851 JXUM01004852 KQ560182 KXJ83967.1 CH916367 EDW01452.1 AXCN02000593 GAKP01008431 JAC50521.1 GDHF01032923 JAI19391.1 GDHF01019067 JAI33247.1 CH477221 EAT47318.1 KU881664 ANC48002.1 GBXI01000372 JAD13920.1 GDHF01001970 JAI50344.1 ADMH02000632 ETN65499.1 GDHF01014150 JAI38164.1 GGFM01006038 MBW26789.1 GGFM01003500 MBW24251.1 GBXI01013719 JAD00573.1 KY011960 AQM52455.1 GGFJ01004884 MBW54025.1 AAZX01006819 GAMC01012391 JAB94164.1 CH902619 KPU76964.1 CH954177 EDV58181.2 KQS70322.1 HACA01032602 CDW49963.1 AE013599 AHN56057.1 EDV37665.2 CH940648 EDW61626.1 KPU76963.1 KRF80119.1 AY190939 AAO00999.1 GGFK01009140 MBW42461.1 BT050472 ACJ13179.1 ACL83084.2 AY051842 GFTR01005775 JAW10651.1 CP012524 ALC42145.1 CH933808 KRG04384.1 JF967750 AEI84584.1 CH480816 EDW47182.1 CM000157 KRJ98547.1 CM002911 KMY92624.1 EDW08980.1 KRJ98548.1 EDW89582.1 CAEY01000026 KQ434777 KZC04171.1 KMY92625.1 QOIP01000002 RLU25558.1 HACA01032603 CDW49964.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000235965
+ More
UP000094527 UP000027135 UP000075882 UP000092445 UP000002320 UP000075920 UP000091820 UP000075883 UP000092460 UP000075902 UP000092443 UP000075900 UP000095301 UP000075885 UP000075903 UP000092444 UP000030765 UP000007062 UP000075840 UP000078200 UP000076408 UP000037069 UP000198287 UP000076407 UP000001554 UP000085678 UP000069940 UP000249989 UP000007798 UP000095300 UP000001070 UP000075886 UP000075884 UP000192221 UP000008820 UP000000673 UP000002358 UP000007801 UP000008711 UP000000803 UP000008792 UP000092553 UP000069272 UP000009192 UP000001292 UP000002282 UP000015104 UP000076502 UP000279307
UP000094527 UP000027135 UP000075882 UP000092445 UP000002320 UP000075920 UP000091820 UP000075883 UP000092460 UP000075902 UP000092443 UP000075900 UP000095301 UP000075885 UP000075903 UP000092444 UP000030765 UP000007062 UP000075840 UP000078200 UP000076408 UP000037069 UP000198287 UP000076407 UP000001554 UP000085678 UP000069940 UP000249989 UP000007798 UP000095300 UP000001070 UP000075886 UP000075884 UP000192221 UP000008820 UP000000673 UP000002358 UP000007801 UP000008711 UP000000803 UP000008792 UP000092553 UP000069272 UP000009192 UP000001292 UP000002282 UP000015104 UP000076502 UP000279307
Interpro
IPR013017
NHL_repeat_subgr
+ More
IPR011042 6-blade_b-propeller_TolB-like
IPR000720 PHM/PAL
IPR001258 NHL_repeat
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR014783 Cu2_ascorb_mOase_CS-2
IPR008977 PHM/PNGase_F_dom_sf
IPR024548 Cu2_monoox_C
IPR020611 Cu2_ascorb_mOase_CS-1
IPR000323 Cu2_ascorb_mOase_N
IPR014784 Cu2_ascorb_mOase-like_C
IPR036939 Cu2_ascorb_mOase_N_sf
IPR011042 6-blade_b-propeller_TolB-like
IPR000720 PHM/PAL
IPR001258 NHL_repeat
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR014783 Cu2_ascorb_mOase_CS-2
IPR008977 PHM/PNGase_F_dom_sf
IPR024548 Cu2_monoox_C
IPR020611 Cu2_ascorb_mOase_CS-1
IPR000323 Cu2_ascorb_mOase_N
IPR014784 Cu2_ascorb_mOase-like_C
IPR036939 Cu2_ascorb_mOase_N_sf
Gene 3D
ProteinModelPortal
H9J8H3
A0A2A4J8Y4
A0A2H1WA59
A0A0N0PBR4
A0A212EKT6
A0A194PQV8
+ More
A0A2J7QPM8 A0A2J7QPM1 A0A1D2NEY4 A0A067RHL7 A0A1Q3FGN2 A0A1Q3FGH4 A0A182KRQ4 A0A1A9ZCM1 B0VZE5 A0A182WIP2 A0A1A9X3N3 T1P8C3 A0A182M7E9 T1PM65 A0A1B0BH65 A0A182TDV8 A0A1A9YAZ3 A0A182RHT7 A0A1I8M5C6 A0A182P3M3 A0A182UW58 A0A1B0GDJ1 A0A084VWY7 Q7QJP6 A0A182IGW0 A0A1A9VP13 A0A182XZ69 A0A0L0BPQ8 A0A226F6T8 A0A1B6LWW9 A0A1B6G878 A0A182X3E7 C3YQ66 A0A1S3I9G3 A0A1S3IAE2 A0A1B6IDE5 A0A182GNQ6 A0A023ESW6 B4MRI5 A0A1I8PLY6 A0A182H9U8 B4J9H1 A0A182QCA2 A0A034W4J0 A0A182NHJ0 A0A0K8TYP3 A0A1W4UZC7 A0A0K8V2X3 Q17KT9 A0A168L575 A0A0A1XSQ6 A0A0K8WH17 W5JRY6 A0A0K8VGX3 A0A1W4UZ23 A0A2M3ZE61 A0A1W4V0N2 A0A2M3Z6W7 A0A0A1WQ78 A0A1Q1NAE3 A0A2M4BLU6 K7IX33 W8BY31 A0A0N8P0F3 B3N6T2 A0A0Q5WLM8 A0A0K2VHL2 A0A0B4LF14 B3MF37 B4LJX2 A0A0P8ZRB0 A0A0Q9WCX2 Q8I1H1 A0A2M4ANV6 B6IDN2 Q9V5E1 A0A224XRL7 A0A0M4EWG8 A0A182FI38 A0A0Q9X7Q6 G8EWC9 B4HMA5 A0A0R1DU55 A0A0J9RA86 B4KNT5 A0A0R1DMP8 B4NX21 T1KEL8 A0A154NX67 A0A0J9TXQ2 A0A3L8E0K9 A0A0K2VHQ9
A0A2J7QPM8 A0A2J7QPM1 A0A1D2NEY4 A0A067RHL7 A0A1Q3FGN2 A0A1Q3FGH4 A0A182KRQ4 A0A1A9ZCM1 B0VZE5 A0A182WIP2 A0A1A9X3N3 T1P8C3 A0A182M7E9 T1PM65 A0A1B0BH65 A0A182TDV8 A0A1A9YAZ3 A0A182RHT7 A0A1I8M5C6 A0A182P3M3 A0A182UW58 A0A1B0GDJ1 A0A084VWY7 Q7QJP6 A0A182IGW0 A0A1A9VP13 A0A182XZ69 A0A0L0BPQ8 A0A226F6T8 A0A1B6LWW9 A0A1B6G878 A0A182X3E7 C3YQ66 A0A1S3I9G3 A0A1S3IAE2 A0A1B6IDE5 A0A182GNQ6 A0A023ESW6 B4MRI5 A0A1I8PLY6 A0A182H9U8 B4J9H1 A0A182QCA2 A0A034W4J0 A0A182NHJ0 A0A0K8TYP3 A0A1W4UZC7 A0A0K8V2X3 Q17KT9 A0A168L575 A0A0A1XSQ6 A0A0K8WH17 W5JRY6 A0A0K8VGX3 A0A1W4UZ23 A0A2M3ZE61 A0A1W4V0N2 A0A2M3Z6W7 A0A0A1WQ78 A0A1Q1NAE3 A0A2M4BLU6 K7IX33 W8BY31 A0A0N8P0F3 B3N6T2 A0A0Q5WLM8 A0A0K2VHL2 A0A0B4LF14 B3MF37 B4LJX2 A0A0P8ZRB0 A0A0Q9WCX2 Q8I1H1 A0A2M4ANV6 B6IDN2 Q9V5E1 A0A224XRL7 A0A0M4EWG8 A0A182FI38 A0A0Q9X7Q6 G8EWC9 B4HMA5 A0A0R1DU55 A0A0J9RA86 B4KNT5 A0A0R1DMP8 B4NX21 T1KEL8 A0A154NX67 A0A0J9TXQ2 A0A3L8E0K9 A0A0K2VHQ9
PDB
5WM0
E-value=1.35006e-61,
Score=599
Ontologies
GO
Topology
Subcellular location
Cell membrane
Confined to cell bodies. With evidence from 3 publications.
SignalP
Position: 1 - 25,
Likelihood: 0.799207
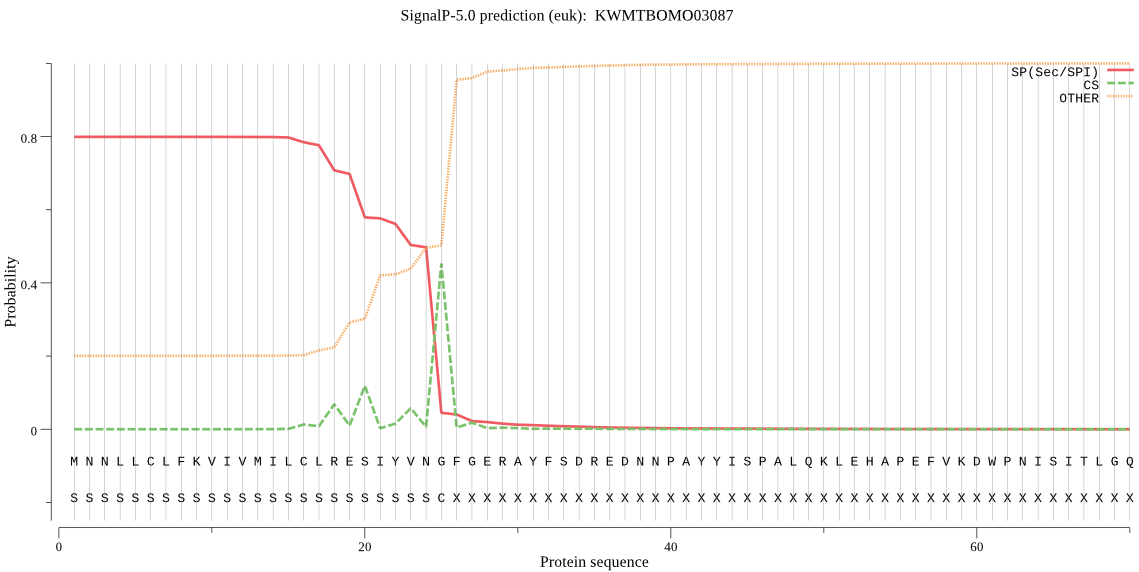
Length:
390
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.999229999999999
Exp number, first 60 AAs:
0.99123
Total prob of N-in:
0.05222
outside
1 - 390
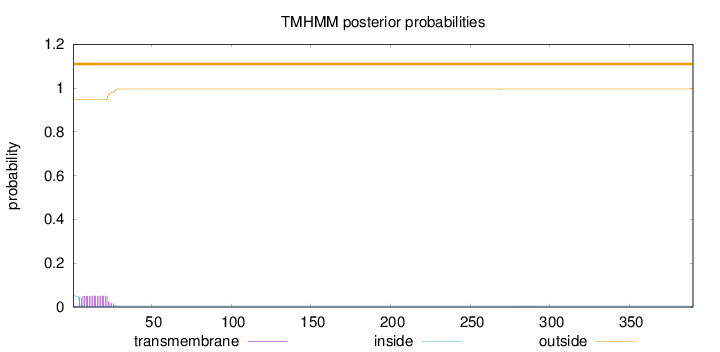
Population Genetic Test Statistics
Pi
220.486825
Theta
165.466216
Tajima's D
1.324054
CLR
0.929944
CSRT
0.741862906854657
Interpretation
Uncertain
