Gene
KWMTBOMO03085
Pre Gene Modal
BGIBMGA005761
Annotation
PREDICTED:_protein_RRNAD1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.076 Mitochondrial Reliability : 1.709 Nuclear Reliability : 1.429
Sequence
CDS
ATGAAGTCTCAGATAAATAACGTAAAAACCTGTATCGATATGTCTCTAAAAGTTATACGTATGTACGATTGGTTGCTGGATTTATATGTATTGGATTTCTTTGTAGACAATCACTGGTATAGGCTTCCGAAATCTTGGCAAGAGAATCTTGATGAGATTGCCTTGGAGCAATTGGGAAATATATTATCAGAAAAACCAACCAATCATATCTTTCCATTGTCATTGCTAGCATTAATTGCCACTGTAAAGGCATTGGGCCTACCACGAAAAAACAGCAAAACTATAAAAAAAGATGTTACACAAATCGCAGACCAATGCAGTACTTATAGCAAAATAAAAAATCTATTCTTGAAACATGTTAAATTGAAGAAGCGGCATGAAGTCAGTGTAATGGCTGAAATCGTTACTGATGTTGCATTGAAATCAAGATGTAATGCAGTTATAGACTTTGGTTCAGGTCTCGGTCATTTAGTGAGGATGTTGGCATATAAGAACAAGTTGTATACTGCAGGCATTGAGTGTCAAAAGCAACTGACGGAGGAGGCTAGAAAACTAGATCTTGAATTTGAATACACTGCAAGAAAATACACCAAAGAAGAGTTTACACCTAATCTACATCGGCCCATACATTATAACATCTCATTATCCAATCATGAGCAACTTCAACATCTGCCTTTACCGACTCATATGACAAACTATGGATTGATTGGGCTCCATCCATGTGGAGATTTAGGCCCTTTACTCTTGAAACACTTCATATGTTCCAATCAGGTGAAATTCATATGTGTTGTAGGCTGTTGTTATACAAAGTTAACAGCTGAAGGATATCCAATAAGCAACTATCTTAAAAAATTAAAAGGTGATCTGTCTTTTGTGAGCAGAGAGATTGCTTGTCACGCTATCGAAATGTACTCTGAGAGATTGAGGCAAGGACTTTATGAAGATTTAAAGGTTCACGCATACAGAGCAGCTCTGGAGAAGATATTAGTTGAACATAATCCCAAATTGAAACACGCTCCAGTTAGGAGTTTGAAACACACCCAAGATTTGAGTTTTGAAAAATACTGTTCGATGGCGGTTGAGAAGCTGTGCTTGCGTTGGCCGCTGCCGGACGTGTCGGAGAGAGACATGTCGGCGCGGGACGAGTCGGTGCGGAGCGTGTCGGCCAGAGACTTGTCGGCGCGGGACGTGTTGGCGTGCGGCGTGTCGGCGCGCGAGGTGTCGGCGGGCGCGGAGGAGGTGCGCGAGTGGCGGCGCGTGGTGGTGGTGTACACGCTGCGGCTGGCGCTGGCGCCGCTGGTCGAGACCGTGCTTTTGTTGGACAGACTGCTCTTTGTGTTGGAACACGGTTTATCCTGTGAGATCCGTCCGATTTTCGACCCGAAGTTGTCTCCAAGGAATCACGTGATAATTGCAAGAAAACCTAATTAG
Protein
MKSQINNVKTCIDMSLKVIRMYDWLLDLYVLDFFVDNHWYRLPKSWQENLDEIALEQLGNILSEKPTNHIFPLSLLALIATVKALGLPRKNSKTIKKDVTQIADQCSTYSKIKNLFLKHVKLKKRHEVSVMAEIVTDVALKSRCNAVIDFGSGLGHLVRMLAYKNKLYTAGIECQKQLTEEARKLDLEFEYTARKYTKEEFTPNLHRPIHYNISLSNHEQLQHLPLPTHMTNYGLIGLHPCGDLGPLLLKHFICSNQVKFICVVGCCYTKLTAEGYPISNYLKKLKGDLSFVSREIACHAIEMYSERLRQGLYEDLKVHAYRAALEKILVEHNPKLKHAPVRSLKHTQDLSFEKYCSMAVEKLCLRWPLPDVSERDMSARDESVRSVSARDLSARDVLACGVSAREVSAGAEEVREWRRVVVVYTLRLALAPLVETVLLLDRLLFVLEHGLSCEIRPIFDPKLSPRNHVIIARKPN
Summary
Uniprot
H9J8B9
A0A2A4JAH1
S4PX59
A0A2H1WAQ6
A0A194PQ87
A0A212EKT2
+ More
A0A2P8XL83 A0A2J7QPN1 A0A067RIL5 A0A1W4WB34 A0A1Y1MKA7 A0A0L0BPU3 A0A2J7QPM4 B3N9B6 A0A0L7R7T7 A0A0J9R7U6 A8DY65 B4QF75 A0A0Q9WGR1 B4LKQ9 B4HRG7 A0A026WY32 B4KRA3 A0A2A3EHN1 A0A1I8M294 B3MGB7 A0A0J9R8Y7 A0A088AJB0 A0A154NWW3 D6W7X6 A0A1L8DGH9 A0A1A9X3N4 A0A195BA52 A0A195EV03 F4WP10 Q290Y9 A0A158NPK6 B4MRL1 A0A151WLZ7 B4P264 A0A1I8Q3N7 W8BM86 A0A0C9QI88 A0A1A9VP14 B7S8Y8 A0A195E585 E2B116 A0A1B0BH64 A0A0Q5WC72 A0A1A9ZCL9 E2B3U7 A0A3B0J622 A0A1A9YAZ4 B7S8Q6 A0A0J9R7L9 A0A1W4VZD8 B4QF73 A0A0J9TVT8 A9UNB1 K7J5J4 A0A3L8DWE3 B4GB15 A0A0Q9WFW3 A0A195CMF2 Q7QJP7 A0A310S7P3 A0A0J9R7U9 A0A182KRQ6 A0A182X3E8 A0A0K8UA55 A0A182I3M1 A0A182VNC2 B4J7V3 A0A182U887 W8B7A2 A0A0M4EDL2 A0A0R1DTB1 S6D4V8 A0A0A1XNS2 G3WTR8 A0A0Q9X882 A0A0P5CFB9 C3Z7E1 A0A182QV05 U4UKN0 E0VQ57 N6TGH2 A0A182M5S5 A0A1B0C8Q7 A0A182WIP3 A0A0K8VVZ6 A0A232FHH5 A0A182P3M2 A0A1S3AIR8 A0A182J217 A0A182RHT8 A0A0P6JC24
A0A2P8XL83 A0A2J7QPN1 A0A067RIL5 A0A1W4WB34 A0A1Y1MKA7 A0A0L0BPU3 A0A2J7QPM4 B3N9B6 A0A0L7R7T7 A0A0J9R7U6 A8DY65 B4QF75 A0A0Q9WGR1 B4LKQ9 B4HRG7 A0A026WY32 B4KRA3 A0A2A3EHN1 A0A1I8M294 B3MGB7 A0A0J9R8Y7 A0A088AJB0 A0A154NWW3 D6W7X6 A0A1L8DGH9 A0A1A9X3N4 A0A195BA52 A0A195EV03 F4WP10 Q290Y9 A0A158NPK6 B4MRL1 A0A151WLZ7 B4P264 A0A1I8Q3N7 W8BM86 A0A0C9QI88 A0A1A9VP14 B7S8Y8 A0A195E585 E2B116 A0A1B0BH64 A0A0Q5WC72 A0A1A9ZCL9 E2B3U7 A0A3B0J622 A0A1A9YAZ4 B7S8Q6 A0A0J9R7L9 A0A1W4VZD8 B4QF73 A0A0J9TVT8 A9UNB1 K7J5J4 A0A3L8DWE3 B4GB15 A0A0Q9WFW3 A0A195CMF2 Q7QJP7 A0A310S7P3 A0A0J9R7U9 A0A182KRQ6 A0A182X3E8 A0A0K8UA55 A0A182I3M1 A0A182VNC2 B4J7V3 A0A182U887 W8B7A2 A0A0M4EDL2 A0A0R1DTB1 S6D4V8 A0A0A1XNS2 G3WTR8 A0A0Q9X882 A0A0P5CFB9 C3Z7E1 A0A182QV05 U4UKN0 E0VQ57 N6TGH2 A0A182M5S5 A0A1B0C8Q7 A0A182WIP3 A0A0K8VVZ6 A0A232FHH5 A0A182P3M2 A0A1S3AIR8 A0A182J217 A0A182RHT8 A0A0P6JC24
Pubmed
19121390
23622113
26354079
22118469
29403074
24845553
+ More
28004739 26108605 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24508170 25315136 18362917 19820115 21719571 15632085 21347285 17550304 24495485 20798317 26109357 26109356 20075255 30249741 12364791 14747013 17210077 20966253 23938757 25830018 21709235 18563158 23537049 20566863 28648823
28004739 26108605 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24508170 25315136 18362917 19820115 21719571 15632085 21347285 17550304 24495485 20798317 26109357 26109356 20075255 30249741 12364791 14747013 17210077 20966253 23938757 25830018 21709235 18563158 23537049 20566863 28648823
EMBL
BABH01027412
NWSH01002324
PCG68524.1
GAIX01005608
JAA86952.1
ODYU01007288
+ More
SOQ49932.1 KQ459597 KPI95128.1 AGBW02014213 OWR42100.1 PYGN01001804 PSN32724.1 NEVH01012088 PNF30535.1 KK852659 KDR19136.1 GEZM01032196 GEZM01032193 JAV84785.1 JRES01001567 KNC21988.1 PNF30534.1 CH954177 EDV59603.2 KQ414637 KOC66940.1 CM002911 KMY92138.1 AE013599 ABV53725.1 CM000362 EDX06104.1 CH940648 KRF80238.1 EDW61782.2 CH480816 EDW46849.1 KK107073 EZA60626.1 CH933808 EDW09319.2 KZ288244 PBC31227.1 CH902619 EDV37820.2 KMY92139.1 KQ434777 KZC04159.1 KQ971307 EFA11151.2 GFDF01008624 JAV05460.1 KQ976542 KYM81074.1 KQ981965 KYN31722.1 GL888243 EGI63941.1 CM000071 EAL25223.3 ADTU01022555 CH963850 EDW74750.2 KQ982944 KYQ48894.1 CM000157 EDW89265.2 GAMC01012094 GAMC01012088 JAB94461.1 GBYB01000262 JAG70029.1 EF710656 ACE75363.1 KQ979608 KYN20350.1 GL444771 EFN60617.1 JXJN01014196 KQS70908.1 GL445407 EFN89616.1 OUUW01000001 SPP75182.1 EF710650 ACE75279.1 KMY92137.1 EDX06102.1 KMY92140.1 BT031275 AAF59163.4 ABY20516.1 QOIP01000003 RLU24757.1 CH479181 EDW32117.1 KRF80239.1 KQ977574 KYN01870.1 AAAB01008807 EAA03936.4 KQ772178 OAD52468.1 KMY92141.1 GDHF01028780 GDHF01027981 JAI23534.1 JAI24333.1 APCN01000215 CH916367 EDW01159.1 GAMC01012093 JAB94462.1 CP012524 ALC42015.1 KRJ98337.1 HF586476 CCQ71313.1 GBXI01001313 JAD12979.1 AEFK01162048 KRG04587.1 GDIP01171134 JAJ52268.1 GG666590 EEN51737.1 AXCN02000593 KB632399 ERL94634.1 DS235389 EEB15513.1 APGK01039164 KB740967 ENN76878.1 AXCM01005430 AJWK01001259 GDHF01009265 JAI43049.1 NNAY01000183 OXU30216.1 GDIQ01009489 JAN85248.1
SOQ49932.1 KQ459597 KPI95128.1 AGBW02014213 OWR42100.1 PYGN01001804 PSN32724.1 NEVH01012088 PNF30535.1 KK852659 KDR19136.1 GEZM01032196 GEZM01032193 JAV84785.1 JRES01001567 KNC21988.1 PNF30534.1 CH954177 EDV59603.2 KQ414637 KOC66940.1 CM002911 KMY92138.1 AE013599 ABV53725.1 CM000362 EDX06104.1 CH940648 KRF80238.1 EDW61782.2 CH480816 EDW46849.1 KK107073 EZA60626.1 CH933808 EDW09319.2 KZ288244 PBC31227.1 CH902619 EDV37820.2 KMY92139.1 KQ434777 KZC04159.1 KQ971307 EFA11151.2 GFDF01008624 JAV05460.1 KQ976542 KYM81074.1 KQ981965 KYN31722.1 GL888243 EGI63941.1 CM000071 EAL25223.3 ADTU01022555 CH963850 EDW74750.2 KQ982944 KYQ48894.1 CM000157 EDW89265.2 GAMC01012094 GAMC01012088 JAB94461.1 GBYB01000262 JAG70029.1 EF710656 ACE75363.1 KQ979608 KYN20350.1 GL444771 EFN60617.1 JXJN01014196 KQS70908.1 GL445407 EFN89616.1 OUUW01000001 SPP75182.1 EF710650 ACE75279.1 KMY92137.1 EDX06102.1 KMY92140.1 BT031275 AAF59163.4 ABY20516.1 QOIP01000003 RLU24757.1 CH479181 EDW32117.1 KRF80239.1 KQ977574 KYN01870.1 AAAB01008807 EAA03936.4 KQ772178 OAD52468.1 KMY92141.1 GDHF01028780 GDHF01027981 JAI23534.1 JAI24333.1 APCN01000215 CH916367 EDW01159.1 GAMC01012093 JAB94462.1 CP012524 ALC42015.1 KRJ98337.1 HF586476 CCQ71313.1 GBXI01001313 JAD12979.1 AEFK01162048 KRG04587.1 GDIP01171134 JAJ52268.1 GG666590 EEN51737.1 AXCN02000593 KB632399 ERL94634.1 DS235389 EEB15513.1 APGK01039164 KB740967 ENN76878.1 AXCM01005430 AJWK01001259 GDHF01009265 JAI43049.1 NNAY01000183 OXU30216.1 GDIQ01009489 JAN85248.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000245037
UP000235965
+ More
UP000027135 UP000192221 UP000037069 UP000008711 UP000053825 UP000000803 UP000000304 UP000008792 UP000001292 UP000053097 UP000009192 UP000242457 UP000095301 UP000007801 UP000005203 UP000076502 UP000007266 UP000091820 UP000078540 UP000078541 UP000007755 UP000001819 UP000005205 UP000007798 UP000075809 UP000002282 UP000095300 UP000078200 UP000078492 UP000000311 UP000092460 UP000092445 UP000008237 UP000268350 UP000092443 UP000002358 UP000279307 UP000008744 UP000078542 UP000007062 UP000075882 UP000076407 UP000075840 UP000075903 UP000001070 UP000075902 UP000092553 UP000007648 UP000001554 UP000075886 UP000030742 UP000009046 UP000019118 UP000075883 UP000092461 UP000075920 UP000215335 UP000075885 UP000079721 UP000075880 UP000075900
UP000027135 UP000192221 UP000037069 UP000008711 UP000053825 UP000000803 UP000000304 UP000008792 UP000001292 UP000053097 UP000009192 UP000242457 UP000095301 UP000007801 UP000005203 UP000076502 UP000007266 UP000091820 UP000078540 UP000078541 UP000007755 UP000001819 UP000005205 UP000007798 UP000075809 UP000002282 UP000095300 UP000078200 UP000078492 UP000000311 UP000092460 UP000092445 UP000008237 UP000268350 UP000092443 UP000002358 UP000279307 UP000008744 UP000078542 UP000007062 UP000075882 UP000076407 UP000075840 UP000075903 UP000001070 UP000075902 UP000092553 UP000007648 UP000001554 UP000075886 UP000030742 UP000009046 UP000019118 UP000075883 UP000092461 UP000075920 UP000215335 UP000075885 UP000079721 UP000075880 UP000075900
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9J8B9
A0A2A4JAH1
S4PX59
A0A2H1WAQ6
A0A194PQ87
A0A212EKT2
+ More
A0A2P8XL83 A0A2J7QPN1 A0A067RIL5 A0A1W4WB34 A0A1Y1MKA7 A0A0L0BPU3 A0A2J7QPM4 B3N9B6 A0A0L7R7T7 A0A0J9R7U6 A8DY65 B4QF75 A0A0Q9WGR1 B4LKQ9 B4HRG7 A0A026WY32 B4KRA3 A0A2A3EHN1 A0A1I8M294 B3MGB7 A0A0J9R8Y7 A0A088AJB0 A0A154NWW3 D6W7X6 A0A1L8DGH9 A0A1A9X3N4 A0A195BA52 A0A195EV03 F4WP10 Q290Y9 A0A158NPK6 B4MRL1 A0A151WLZ7 B4P264 A0A1I8Q3N7 W8BM86 A0A0C9QI88 A0A1A9VP14 B7S8Y8 A0A195E585 E2B116 A0A1B0BH64 A0A0Q5WC72 A0A1A9ZCL9 E2B3U7 A0A3B0J622 A0A1A9YAZ4 B7S8Q6 A0A0J9R7L9 A0A1W4VZD8 B4QF73 A0A0J9TVT8 A9UNB1 K7J5J4 A0A3L8DWE3 B4GB15 A0A0Q9WFW3 A0A195CMF2 Q7QJP7 A0A310S7P3 A0A0J9R7U9 A0A182KRQ6 A0A182X3E8 A0A0K8UA55 A0A182I3M1 A0A182VNC2 B4J7V3 A0A182U887 W8B7A2 A0A0M4EDL2 A0A0R1DTB1 S6D4V8 A0A0A1XNS2 G3WTR8 A0A0Q9X882 A0A0P5CFB9 C3Z7E1 A0A182QV05 U4UKN0 E0VQ57 N6TGH2 A0A182M5S5 A0A1B0C8Q7 A0A182WIP3 A0A0K8VVZ6 A0A232FHH5 A0A182P3M2 A0A1S3AIR8 A0A182J217 A0A182RHT8 A0A0P6JC24
A0A2P8XL83 A0A2J7QPN1 A0A067RIL5 A0A1W4WB34 A0A1Y1MKA7 A0A0L0BPU3 A0A2J7QPM4 B3N9B6 A0A0L7R7T7 A0A0J9R7U6 A8DY65 B4QF75 A0A0Q9WGR1 B4LKQ9 B4HRG7 A0A026WY32 B4KRA3 A0A2A3EHN1 A0A1I8M294 B3MGB7 A0A0J9R8Y7 A0A088AJB0 A0A154NWW3 D6W7X6 A0A1L8DGH9 A0A1A9X3N4 A0A195BA52 A0A195EV03 F4WP10 Q290Y9 A0A158NPK6 B4MRL1 A0A151WLZ7 B4P264 A0A1I8Q3N7 W8BM86 A0A0C9QI88 A0A1A9VP14 B7S8Y8 A0A195E585 E2B116 A0A1B0BH64 A0A0Q5WC72 A0A1A9ZCL9 E2B3U7 A0A3B0J622 A0A1A9YAZ4 B7S8Q6 A0A0J9R7L9 A0A1W4VZD8 B4QF73 A0A0J9TVT8 A9UNB1 K7J5J4 A0A3L8DWE3 B4GB15 A0A0Q9WFW3 A0A195CMF2 Q7QJP7 A0A310S7P3 A0A0J9R7U9 A0A182KRQ6 A0A182X3E8 A0A0K8UA55 A0A182I3M1 A0A182VNC2 B4J7V3 A0A182U887 W8B7A2 A0A0M4EDL2 A0A0R1DTB1 S6D4V8 A0A0A1XNS2 G3WTR8 A0A0Q9X882 A0A0P5CFB9 C3Z7E1 A0A182QV05 U4UKN0 E0VQ57 N6TGH2 A0A182M5S5 A0A1B0C8Q7 A0A182WIP3 A0A0K8VVZ6 A0A232FHH5 A0A182P3M2 A0A1S3AIR8 A0A182J217 A0A182RHT8 A0A0P6JC24
Ontologies
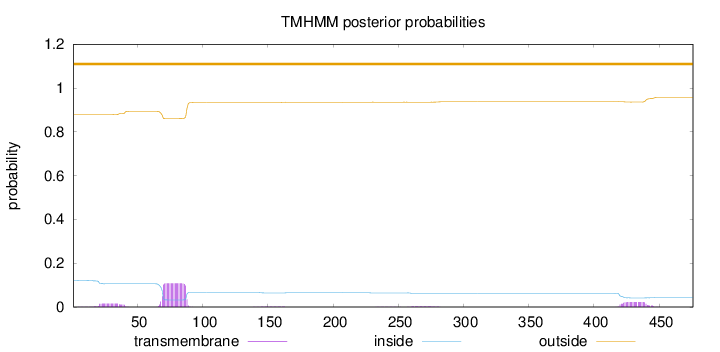
Topology
Length:
476
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.93401
Exp number, first 60 AAs:
0.31254
Total prob of N-in:
0.12193
outside
1 - 476
Population Genetic Test Statistics
Pi
17.743589
Theta
22.407546
Tajima's D
-0.695717
CLR
0.003351
CSRT
0.1976401179941
Interpretation
Uncertain
