Pre Gene Modal
BGIBMGA005816
Annotation
PREDICTED:_mitochondrial_intermediate_peptidase_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.331 Mitochondrial Reliability : 1.743 Nuclear Reliability : 1.043
Sequence
CDS
ATGTCTACAATGAAGTTATTGAAACCCCTCTGGATCCTCCGAAATGGCAGAAAGAAGAAATTGGGATGCTCTGTCAGTACTTGGTCACCTTTGGCAACAGCATTCAACACCAGACCAAATTCCAGACCTATTTTTGACTCTTTAAAGGAAAGAACTGGCCTCTTCAATAAACCAGAGCTCACAACTTTTGAAGGCTTCTACACGCTCAAGGACCAGGCTATCGAAGCAACTGATCGTCTGATAGAAGAGGCCACCAACAGCCCCACGCGTCCTATGGTCGAGATATTTGATGAGCTTTCCGACACACTGTGCAAAGTTGCAGATCTTGCTGAGTTTGTGAGGATAGCTCACCCCCAACCACATTTCGCACGAGCTGCTGAAGAAGCCTGTATCAGCATTAGTGGAGTTGTCGAAAAGTTAAACACGCACAAAGGCTTGTACGAAGCTTTAAAGAAGTCAGTTGAGAATGGAACATCTGGAGACAAACATTTAGCAGAATTATTCTTGTTCGACTTTGAACAGAGCGGAATACACCTCGAAGAGAACAAAAGGAAGAGTGTGGTGGCTCTCAATGATGTTATCCTCCAAACTGGGCAGAGGTTCATGGCCGGAGCTGCTAAACCAAGACGTGTTTCTCGAAGTGCTGTACCTCAGAATGTTAGACATTTCTTCAGCACCGAAGGTGACAATATAATAGTGAGCGGGGTGTACGCGGAGTGCGGGGAGGCGGCCGCCCGCGAGGCTGCCTACCGACTGTACCTAGCGCCGGACGGCACGCAGGACCGCCTGCTGGACGCCGTGCTGCAGGCCAGGCACCACATGGCCACGCTCTGCGGCTTCCAGACCTACGCCGACAGAGCAATAAAAGCGAGTACAATGGAGACAGCAGCAAACGTGCGGCAGTTTCTCGACCTCCTCTCTGACGGCCTCCACGACCGCGCTAGAATGGACTTCGACGTGATGCTGAAGATGAAGAAACAGGAAACGCCATATCTGGACCAATTAAACTGCTGGGACACGCCGTACTATACTCACAAGGCGAAGAACTTGTTATTGGACCAGGCCAATTCGGACTTCAGTCCGTACTTCAGTCTGGGCGCGTGCATGGAGGGCTTGGATATGTTGTGTCAAGAGCTATACGGCATCAGTCTGAAGTCCGAGGAAATGTTGCCAGGCGAGTCGTGGTCTCCGGACGTATACAAGCTGGCCGTGGTCCACGAGTCGGAGGGGCTGCTGGGCCACATCTACTGCGACCTGTACGAGCGGCCCGGGAAGCCGCATCAGGACTGCCACTTCACTATACAGGGCGGGAAGCTGTTACCAGATGGCTCCTACCAGAACCCGATCGTGGTGATAATGTTGTCGCTGAGCGGCGGGCACCGCTCGGGGCCGGCGCTGCTGTCCGCGGGCGCGCTGGACAACCTGTTCCACGAGGCGGGCCACGCGCTGCACTCCATGCTGGGCCGCGCGCCCCACCAGCACGTGGCCGGCACGCGCTGCCCCACCGACCTGGCCGAGCTGCCCAGCGTGCTCATGGAGTGCTTCGCGGCCGACCCGCAGGTGCTGCGCAGGTTCGCGCGGCACTTCCAGACGCGGCAGGCGGCGCCGGAGCGCACGCTGCAGCGCCTGTGCGCCTCCAAGCGACTGTTCGCCGCCAGCGAGATGCAGCTGCAGGTGTTTTATTCAGCCCTGGACCAGCAGTACCACGACCGGCCCGCAGCGCCCGGGGGCCGCACCACGGACGATCTGAAAGAAGTCCAGAAACAATATTATGGACTTCCGTATGTCGAAAACACAGCGTGGCAGCACCGGTTCAGCCACCTGGTCGGCTACGGCGCCAAGTACTACTCCTACCTCATCTCGCGCGCGCTCGCCTCCTCCGCCTGGAACACGCACTTCGCGGCCGACCCGCTCGCGCGCCGCGCCGGCCGCCGCCTGCGGGACCGCGTGCTGCGACACGGGGGCGCCGTGCCGCCGCAGGTCCTGCTCAAGGACTACCTGGAAATGGAGATAACTCCACAGACGCTCGCGTCGGCGCTGATAGACGAGCTGGACTGCCACAAGGACCACCTCGACACCGTGCTAAAGATACTGGACAGAAAGTAA
Protein
MSTMKLLKPLWILRNGRKKKLGCSVSTWSPLATAFNTRPNSRPIFDSLKERTGLFNKPELTTFEGFYTLKDQAIEATDRLIEEATNSPTRPMVEIFDELSDTLCKVADLAEFVRIAHPQPHFARAAEEACISISGVVEKLNTHKGLYEALKKSVENGTSGDKHLAELFLFDFEQSGIHLEENKRKSVVALNDVILQTGQRFMAGAAKPRRVSRSAVPQNVRHFFSTEGDNIIVSGVYAECGEAAAREAAYRLYLAPDGTQDRLLDAVLQARHHMATLCGFQTYADRAIKASTMETAANVRQFLDLLSDGLHDRARMDFDVMLKMKKQETPYLDQLNCWDTPYYTHKAKNLLLDQANSDFSPYFSLGACMEGLDMLCQELYGISLKSEEMLPGESWSPDVYKLAVVHESEGLLGHIYCDLYERPGKPHQDCHFTIQGGKLLPDGSYQNPIVVIMLSLSGGHRSGPALLSAGALDNLFHEAGHALHSMLGRAPHQHVAGTRCPTDLAELPSVLMECFAADPQVLRRFARHFQTRQAAPERTLQRLCASKRLFAASEMQLQVFYSALDQQYHDRPAAPGGRTTDDLKEVQKQYYGLPYVENTAWQHRFSHLVGYGAKYYSYLISRALASSAWNTHFAADPLARRAGRRLRDRVLRHGGAVPPQVLLKDYLEMEITPQTLASALIDELDCHKDHLDTVLKILDRK
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M3 family.
Uniprot
H9J8H4
A0A194PQV3
A0A2H1V1U6
A0A2A4K8W3
A0A212EKU1
A0A1S4F4W5
+ More
Q17F93 A0A084VF33 A0A182J0C4 B0WT45 A0A1Q3F2A6 A0A182QJR8 A0A2M4AJY9 W5JFN9 A0A182FFY8 A0A2M4BFQ7 A0A2M4BFZ3 A0A2M4BFP4 A0A182SPU2 A0A2J7QC24 A0A182WIK2 Q7QJY5 A0A1B0ESP3 A0A182XZI8 A0A182JT41 A0A182R4R0 A0A1L8E140 A0A1L8E193 A0A1L8E1N0 A0A182LRM5 A0A067RKH4 Q28YP7 A0A182U055 A0A1B6C972 A0A1B6HVG2 A0A182LGT9 A0A182I3Y3 B4H8C5 A0A1B6HIE8 A0A3B0JQ62 A0A182USY3 W8BQ27 B4NMV8 A0A0L0BTW9 A0A0K8W4F5 A0A1L8E0X4 A0A0K8UCU5 A0A034WF14 A0A1I8MYY9 T1P7J4 A0A0M4ECE2 A0A1W4VYC8 A0A0M4EEH6 B4J5Q8 B4LNM6 B3MBQ0 B4KM63 A0A2J7QC18 A0A0J9R6B8 B4II08 A0A195FNY2 B4QCP2 A0A1J1I5C7 A0A1I8PML9 A1Z6H6 Q8T8Q1 B3N3I6 A0A2J7QC34 A0A195DWF9 A0A3B0JKC6 A0A151XD88 A0A151IDL4 A0A195B6M0 B4NYX7 A0A2M3ZAS8 A0A1A9XVG3 A0A1A9ZYC7 A0A0A9WMI4 A0A1B0G3G5 F4X128 A0A1B0AXD2 A0A023F246 A0A336JW82 A0A1A9WMX6 U4UE67 A0A3L8DRL2 N6TZD0 E9J3A6 A0A069DWS0 K7J8I2 E2BWA5 A0A232FCB9 A0A1A9V2E5 A0A026WZ09 A0A0P4VQH3 T1HRH4 A0A182XRT2 A0A1L8E109 A0A0V0GCF7
Q17F93 A0A084VF33 A0A182J0C4 B0WT45 A0A1Q3F2A6 A0A182QJR8 A0A2M4AJY9 W5JFN9 A0A182FFY8 A0A2M4BFQ7 A0A2M4BFZ3 A0A2M4BFP4 A0A182SPU2 A0A2J7QC24 A0A182WIK2 Q7QJY5 A0A1B0ESP3 A0A182XZI8 A0A182JT41 A0A182R4R0 A0A1L8E140 A0A1L8E193 A0A1L8E1N0 A0A182LRM5 A0A067RKH4 Q28YP7 A0A182U055 A0A1B6C972 A0A1B6HVG2 A0A182LGT9 A0A182I3Y3 B4H8C5 A0A1B6HIE8 A0A3B0JQ62 A0A182USY3 W8BQ27 B4NMV8 A0A0L0BTW9 A0A0K8W4F5 A0A1L8E0X4 A0A0K8UCU5 A0A034WF14 A0A1I8MYY9 T1P7J4 A0A0M4ECE2 A0A1W4VYC8 A0A0M4EEH6 B4J5Q8 B4LNM6 B3MBQ0 B4KM63 A0A2J7QC18 A0A0J9R6B8 B4II08 A0A195FNY2 B4QCP2 A0A1J1I5C7 A0A1I8PML9 A1Z6H6 Q8T8Q1 B3N3I6 A0A2J7QC34 A0A195DWF9 A0A3B0JKC6 A0A151XD88 A0A151IDL4 A0A195B6M0 B4NYX7 A0A2M3ZAS8 A0A1A9XVG3 A0A1A9ZYC7 A0A0A9WMI4 A0A1B0G3G5 F4X128 A0A1B0AXD2 A0A023F246 A0A336JW82 A0A1A9WMX6 U4UE67 A0A3L8DRL2 N6TZD0 E9J3A6 A0A069DWS0 K7J8I2 E2BWA5 A0A232FCB9 A0A1A9V2E5 A0A026WZ09 A0A0P4VQH3 T1HRH4 A0A182XRT2 A0A1L8E109 A0A0V0GCF7
Pubmed
19121390
26354079
22118469
17510324
24438588
20920257
+ More
23761445 12364791 14747013 17210077 25244985 24845553 15632085 17994087 20966253 24495485 26108605 25348373 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 25401762 21719571 25474469 23537049 30249741 21282665 26334808 20075255 20798317 28648823 24508170 27129103
23761445 12364791 14747013 17210077 25244985 24845553 15632085 17994087 20966253 24495485 26108605 25348373 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 25401762 21719571 25474469 23537049 30249741 21282665 26334808 20075255 20798317 28648823 24508170 27129103
EMBL
BABH01027415
BABH01027416
KQ459597
KPI95124.1
ODYU01000109
SOQ34322.1
+ More
NWSH01000041 PCG80344.1 AGBW02014213 OWR42103.1 CH477272 EAT45254.1 ATLV01012360 KE524785 KFB36577.1 DS232080 EDS34188.1 GFDL01013350 JAV21695.1 AXCN02000894 GGFK01007778 MBW41099.1 ADMH02001644 ETN61629.1 GGFJ01002731 MBW51872.1 GGFJ01002730 MBW51871.1 GGFJ01002729 MBW51870.1 NEVH01016292 PNF26131.1 AAAB01008807 EAA04703.3 AJWK01004522 GFDF01001667 JAV12417.1 GFDF01001668 JAV12416.1 GFDF01001669 JAV12415.1 AXCM01005238 AXCM01005239 AXCM01005240 KK852415 KDR24371.1 CM000071 EAL25918.2 GEDC01027252 JAS10046.1 GECU01029045 JAS78661.1 APCN01000228 CH479223 EDW34960.1 GECU01033223 JAS74483.1 OUUW01000001 SPP75809.1 GAMC01005448 JAC01108.1 CH964282 EDW85697.1 JRES01001351 KNC23456.1 GDHF01022435 GDHF01006328 JAI29879.1 JAI45986.1 GFDF01001691 JAV12393.1 GDHF01027921 JAI24393.1 GAKP01006579 JAC52373.1 KA644442 AFP59071.1 CP012524 ALC41377.1 ALC40435.1 CH916367 EDW01834.1 CH940648 EDW62206.1 CH902619 EDV38196.1 CH933808 EDW08727.2 PNF26136.1 CM002911 KMY91653.1 CH480841 EDW49534.1 KQ981387 KYN42022.1 CM000362 EDX05831.1 CVRI01000042 CRK95543.1 AE013599 AAF57312.1 AY075581 AAL68386.1 CH954177 EDV59868.1 PNF26132.1 KQ980204 KYN17245.1 SPP75810.1 KQ982294 KYQ58290.1 KQ977985 KYM98339.1 KQ976574 KYM80151.1 CM000157 EDW89828.1 GGFM01004895 MBW25646.1 GBHO01033947 GBRD01013110 JAG09657.1 JAG52716.1 CCAG010013345 GL888525 EGI59839.1 JXJN01005228 GBBI01003294 JAC15418.1 UFQS01000006 UFQT01000006 SSW97016.1 SSX17403.1 KB632254 ERL90623.1 QOIP01000005 RLU23085.1 APGK01055188 KB741261 ENN71597.1 GL768061 EFZ12674.1 GBGD01000737 JAC88152.1 GL451103 EFN80009.1 NNAY01000478 OXU28113.1 KK107063 EZA60986.1 GDKW01001885 JAI54710.1 ACPB03004826 GFDF01001697 JAV12387.1 GECL01001053 JAP05071.1
NWSH01000041 PCG80344.1 AGBW02014213 OWR42103.1 CH477272 EAT45254.1 ATLV01012360 KE524785 KFB36577.1 DS232080 EDS34188.1 GFDL01013350 JAV21695.1 AXCN02000894 GGFK01007778 MBW41099.1 ADMH02001644 ETN61629.1 GGFJ01002731 MBW51872.1 GGFJ01002730 MBW51871.1 GGFJ01002729 MBW51870.1 NEVH01016292 PNF26131.1 AAAB01008807 EAA04703.3 AJWK01004522 GFDF01001667 JAV12417.1 GFDF01001668 JAV12416.1 GFDF01001669 JAV12415.1 AXCM01005238 AXCM01005239 AXCM01005240 KK852415 KDR24371.1 CM000071 EAL25918.2 GEDC01027252 JAS10046.1 GECU01029045 JAS78661.1 APCN01000228 CH479223 EDW34960.1 GECU01033223 JAS74483.1 OUUW01000001 SPP75809.1 GAMC01005448 JAC01108.1 CH964282 EDW85697.1 JRES01001351 KNC23456.1 GDHF01022435 GDHF01006328 JAI29879.1 JAI45986.1 GFDF01001691 JAV12393.1 GDHF01027921 JAI24393.1 GAKP01006579 JAC52373.1 KA644442 AFP59071.1 CP012524 ALC41377.1 ALC40435.1 CH916367 EDW01834.1 CH940648 EDW62206.1 CH902619 EDV38196.1 CH933808 EDW08727.2 PNF26136.1 CM002911 KMY91653.1 CH480841 EDW49534.1 KQ981387 KYN42022.1 CM000362 EDX05831.1 CVRI01000042 CRK95543.1 AE013599 AAF57312.1 AY075581 AAL68386.1 CH954177 EDV59868.1 PNF26132.1 KQ980204 KYN17245.1 SPP75810.1 KQ982294 KYQ58290.1 KQ977985 KYM98339.1 KQ976574 KYM80151.1 CM000157 EDW89828.1 GGFM01004895 MBW25646.1 GBHO01033947 GBRD01013110 JAG09657.1 JAG52716.1 CCAG010013345 GL888525 EGI59839.1 JXJN01005228 GBBI01003294 JAC15418.1 UFQS01000006 UFQT01000006 SSW97016.1 SSX17403.1 KB632254 ERL90623.1 QOIP01000005 RLU23085.1 APGK01055188 KB741261 ENN71597.1 GL768061 EFZ12674.1 GBGD01000737 JAC88152.1 GL451103 EFN80009.1 NNAY01000478 OXU28113.1 KK107063 EZA60986.1 GDKW01001885 JAI54710.1 ACPB03004826 GFDF01001697 JAV12387.1 GECL01001053 JAP05071.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000008820
UP000030765
+ More
UP000075880 UP000002320 UP000075886 UP000000673 UP000069272 UP000075901 UP000235965 UP000075920 UP000007062 UP000092461 UP000076408 UP000075881 UP000075900 UP000075883 UP000027135 UP000001819 UP000075902 UP000075882 UP000075840 UP000008744 UP000268350 UP000075903 UP000007798 UP000037069 UP000095301 UP000092553 UP000192221 UP000001070 UP000008792 UP000007801 UP000009192 UP000001292 UP000078541 UP000000304 UP000183832 UP000095300 UP000000803 UP000008711 UP000078492 UP000075809 UP000078542 UP000078540 UP000002282 UP000092443 UP000092445 UP000092444 UP000007755 UP000092460 UP000091820 UP000030742 UP000279307 UP000019118 UP000002358 UP000008237 UP000215335 UP000078200 UP000053097 UP000015103 UP000076407
UP000075880 UP000002320 UP000075886 UP000000673 UP000069272 UP000075901 UP000235965 UP000075920 UP000007062 UP000092461 UP000076408 UP000075881 UP000075900 UP000075883 UP000027135 UP000001819 UP000075902 UP000075882 UP000075840 UP000008744 UP000268350 UP000075903 UP000007798 UP000037069 UP000095301 UP000092553 UP000192221 UP000001070 UP000008792 UP000007801 UP000009192 UP000001292 UP000078541 UP000000304 UP000183832 UP000095300 UP000000803 UP000008711 UP000078492 UP000075809 UP000078542 UP000078540 UP000002282 UP000092443 UP000092445 UP000092444 UP000007755 UP000092460 UP000091820 UP000030742 UP000279307 UP000019118 UP000002358 UP000008237 UP000215335 UP000078200 UP000053097 UP000015103 UP000076407
Interpro
IPR001567
Pept_M3A_M3B
+ More
IPR024077 Neurolysin/TOP_dom2
IPR033851 M3A_MIP
IPR024079 MetalloPept_cat_dom_sf
IPR015919 Cadherin-like_sf
IPR002126 Cadherin-like_dom
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR037289 Elp2
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR006594 LisH
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR024077 Neurolysin/TOP_dom2
IPR033851 M3A_MIP
IPR024079 MetalloPept_cat_dom_sf
IPR015919 Cadherin-like_sf
IPR002126 Cadherin-like_dom
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR037289 Elp2
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR006594 LisH
IPR011989 ARM-like
IPR016024 ARM-type_fold
Gene 3D
CDD
ProteinModelPortal
H9J8H4
A0A194PQV3
A0A2H1V1U6
A0A2A4K8W3
A0A212EKU1
A0A1S4F4W5
+ More
Q17F93 A0A084VF33 A0A182J0C4 B0WT45 A0A1Q3F2A6 A0A182QJR8 A0A2M4AJY9 W5JFN9 A0A182FFY8 A0A2M4BFQ7 A0A2M4BFZ3 A0A2M4BFP4 A0A182SPU2 A0A2J7QC24 A0A182WIK2 Q7QJY5 A0A1B0ESP3 A0A182XZI8 A0A182JT41 A0A182R4R0 A0A1L8E140 A0A1L8E193 A0A1L8E1N0 A0A182LRM5 A0A067RKH4 Q28YP7 A0A182U055 A0A1B6C972 A0A1B6HVG2 A0A182LGT9 A0A182I3Y3 B4H8C5 A0A1B6HIE8 A0A3B0JQ62 A0A182USY3 W8BQ27 B4NMV8 A0A0L0BTW9 A0A0K8W4F5 A0A1L8E0X4 A0A0K8UCU5 A0A034WF14 A0A1I8MYY9 T1P7J4 A0A0M4ECE2 A0A1W4VYC8 A0A0M4EEH6 B4J5Q8 B4LNM6 B3MBQ0 B4KM63 A0A2J7QC18 A0A0J9R6B8 B4II08 A0A195FNY2 B4QCP2 A0A1J1I5C7 A0A1I8PML9 A1Z6H6 Q8T8Q1 B3N3I6 A0A2J7QC34 A0A195DWF9 A0A3B0JKC6 A0A151XD88 A0A151IDL4 A0A195B6M0 B4NYX7 A0A2M3ZAS8 A0A1A9XVG3 A0A1A9ZYC7 A0A0A9WMI4 A0A1B0G3G5 F4X128 A0A1B0AXD2 A0A023F246 A0A336JW82 A0A1A9WMX6 U4UE67 A0A3L8DRL2 N6TZD0 E9J3A6 A0A069DWS0 K7J8I2 E2BWA5 A0A232FCB9 A0A1A9V2E5 A0A026WZ09 A0A0P4VQH3 T1HRH4 A0A182XRT2 A0A1L8E109 A0A0V0GCF7
Q17F93 A0A084VF33 A0A182J0C4 B0WT45 A0A1Q3F2A6 A0A182QJR8 A0A2M4AJY9 W5JFN9 A0A182FFY8 A0A2M4BFQ7 A0A2M4BFZ3 A0A2M4BFP4 A0A182SPU2 A0A2J7QC24 A0A182WIK2 Q7QJY5 A0A1B0ESP3 A0A182XZI8 A0A182JT41 A0A182R4R0 A0A1L8E140 A0A1L8E193 A0A1L8E1N0 A0A182LRM5 A0A067RKH4 Q28YP7 A0A182U055 A0A1B6C972 A0A1B6HVG2 A0A182LGT9 A0A182I3Y3 B4H8C5 A0A1B6HIE8 A0A3B0JQ62 A0A182USY3 W8BQ27 B4NMV8 A0A0L0BTW9 A0A0K8W4F5 A0A1L8E0X4 A0A0K8UCU5 A0A034WF14 A0A1I8MYY9 T1P7J4 A0A0M4ECE2 A0A1W4VYC8 A0A0M4EEH6 B4J5Q8 B4LNM6 B3MBQ0 B4KM63 A0A2J7QC18 A0A0J9R6B8 B4II08 A0A195FNY2 B4QCP2 A0A1J1I5C7 A0A1I8PML9 A1Z6H6 Q8T8Q1 B3N3I6 A0A2J7QC34 A0A195DWF9 A0A3B0JKC6 A0A151XD88 A0A151IDL4 A0A195B6M0 B4NYX7 A0A2M3ZAS8 A0A1A9XVG3 A0A1A9ZYC7 A0A0A9WMI4 A0A1B0G3G5 F4X128 A0A1B0AXD2 A0A023F246 A0A336JW82 A0A1A9WMX6 U4UE67 A0A3L8DRL2 N6TZD0 E9J3A6 A0A069DWS0 K7J8I2 E2BWA5 A0A232FCB9 A0A1A9V2E5 A0A026WZ09 A0A0P4VQH3 T1HRH4 A0A182XRT2 A0A1L8E109 A0A0V0GCF7
PDB
2O36
E-value=3.61546e-36,
Score=382
Ontologies
GO
PANTHER
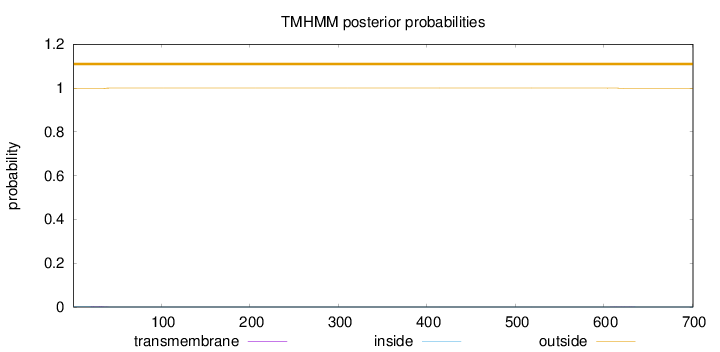
Topology
Length:
701
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03442
Exp number, first 60 AAs:
0.0265
Total prob of N-in:
0.00159
outside
1 - 701
Population Genetic Test Statistics
Pi
251.416502
Theta
170.165751
Tajima's D
1.762067
CLR
0.005793
CSRT
0.84030798460077
Interpretation
Uncertain
