Gene
KWMTBOMO03079
Pre Gene Modal
BGIBMGA005817
Annotation
PREDICTED:_aquaporin-2-like_[Papilio_xuthus]
Full name
Neurogenic protein big brain
Location in the cell
PlasmaMembrane Reliability : 2.397
Sequence
CDS
ATGGGCGAACAAACGTTTACCATACCAGACGATAATCTGGAACAGCACATTGTGACTTTGTTTGAGAAATTGGAAATTTTACGCCGGGACGCGAATACGAATGGGGACAATTTGGCGGGCAGGATTCCAGCTAGATTAGAGCTGAGGACTTTGTCCCTCTGGAAGGCCGTGGTCGCAGAATGTGTGGCGTCGTTCCTTTATGTTTTCATAGTGTGCGGGGCGGCTGGTGGGGCGGGTGTGGGCGCCTCAGCCTCAGCAGTCCTTCTATCTACGGCACTGGCTTCAGGATGTGCTATAGCAACCCTCACCATGTGCTTCGTTAATATCTCTGGTGCTCACGTGAACCCGGCAGTATCGGCAGCACTGTGCATCCGAAGACACGTGTCTCCGCTCCGAGCCGCGTTGTACGTCATGGCACAGTGCGGAGGTGCGATCGCAGGAGCGGCCGCCATTTACGGGTAA
Protein
MGEQTFTIPDDNLEQHIVTLFEKLEILRRDANTNGDNLAGRIPARLELRTLSLWKAVVAECVASFLYVFIVCGAAGGAGVGASASAVLLSTALASGCAIATLTMCFVNISGAHVNPAVSAALCIRRHVSPLRAALYVMAQCGGAIAGAAAIYG
Summary
Description
Essential for proper differentiation of ectoderm. Acts synergistically with neurogenic locus proteins Notch and Delta during the separation of neural and epidermal cell lineages in response to the lateral inhibition signal. Voltage-insensitive monovalent cation channel. Ion transport is blocked by the presence of divalent cations.
Miscellaneous
Separation of neuroblasts from the ectoderm into the inner part of embryo is one of the first steps of CNS development in insects, this process is under control of the neurogenic genes. Mutation in bib gene underlies "big brain" development defect.
Similarity
Belongs to the MIP/aquaporin (TC 1.A.8) family.
Keywords
Complete proteome
Developmental protein
Differentiation
Ion channel
Ion transport
Membrane
Neurogenesis
Phosphoprotein
Reference proteome
Repeat
Transmembrane
Transmembrane helix
Transport
Feature
chain Neurogenic protein big brain
Uniprot
A0A2H1VDQ3
A0A194PP89
A0A0N1IEB0
A0A2A4K7Y4
A0A212EKV3
H9J8H5
+ More
A0A0L7LBI9 A0A1W4USY7 B4Q8B5 P23645 B4NYR0 M9PB86 B3N909 A0A0J9QZM1 A0A1B0BJI5 A0A1A9YA33 A0A3B0JU36 A0A1I8Q664 A0A1I8M1G5 A0A1B0D6Y8 L7P705 B4G7T7 B0XAC5 W5J377 Q29NF9 B4M9F0 A0A1A9WW00 B4HWE5 B4JCH1 A0A0L0CGT1 O17044 B4KH40 B3MPC6 A0A0M4EC41 Q17BX1 A0A1I8JSJ9 A0A182JXF3 A0A034VYC6 A0A0M4E804 A0A2K6VB93 A0A182M7A1 A0A0K8UVA6 W8C2H2 A0A182S9X3 A0A182TUN2 A0A182KWM9 Q7Q873 A0A084VMQ5 A0A240PMK8 A0A3F2YW06 A0A1Y9IVN1 A0A1Y9GKD2 A0A1I8JU95 A0A168SE93 A0A1S4GZW8 A0A240PPM5 B4MX36 A0A182Y8J3 A0A182QNB7 A0A0T6ATC6 A0A182VKX8 A0A336M7F0 A0A1Y9J072 A0A182X4Z5 A0A182Y8J1 A0A182MBR0 A0A182IT42 A0A182UCW2 A0A165Y505 A0A067R979 A0A146LC65 A0A182NZ12 A0A0A9XE43 A0A0A9XMC6 A0A146LG91 Q7Q872 A0A084VMQ6 A0A182FZQ0 A0A182PYX1 A0A182II08 A0A1S4GZY8 A0A182VC62 A0A182WQK0 A0A182KWN1 A0A182S4K2 A0A182R1E8 D6W7H3 E0W4C7 A0A346A6C0 A0A0B6VQ56 U4UMC4 A0A0P6IWW3 A0A0P4WPZ7 A0A1J1ILL8 N6U0L5 A0A165AE12 A0A0P6FHE8 A0A0P4WBD5 A0A2P6KTE2 E9GCI3
A0A0L7LBI9 A0A1W4USY7 B4Q8B5 P23645 B4NYR0 M9PB86 B3N909 A0A0J9QZM1 A0A1B0BJI5 A0A1A9YA33 A0A3B0JU36 A0A1I8Q664 A0A1I8M1G5 A0A1B0D6Y8 L7P705 B4G7T7 B0XAC5 W5J377 Q29NF9 B4M9F0 A0A1A9WW00 B4HWE5 B4JCH1 A0A0L0CGT1 O17044 B4KH40 B3MPC6 A0A0M4EC41 Q17BX1 A0A1I8JSJ9 A0A182JXF3 A0A034VYC6 A0A0M4E804 A0A2K6VB93 A0A182M7A1 A0A0K8UVA6 W8C2H2 A0A182S9X3 A0A182TUN2 A0A182KWM9 Q7Q873 A0A084VMQ5 A0A240PMK8 A0A3F2YW06 A0A1Y9IVN1 A0A1Y9GKD2 A0A1I8JU95 A0A168SE93 A0A1S4GZW8 A0A240PPM5 B4MX36 A0A182Y8J3 A0A182QNB7 A0A0T6ATC6 A0A182VKX8 A0A336M7F0 A0A1Y9J072 A0A182X4Z5 A0A182Y8J1 A0A182MBR0 A0A182IT42 A0A182UCW2 A0A165Y505 A0A067R979 A0A146LC65 A0A182NZ12 A0A0A9XE43 A0A0A9XMC6 A0A146LG91 Q7Q872 A0A084VMQ6 A0A182FZQ0 A0A182PYX1 A0A182II08 A0A1S4GZY8 A0A182VC62 A0A182WQK0 A0A182KWN1 A0A182S4K2 A0A182R1E8 D6W7H3 E0W4C7 A0A346A6C0 A0A0B6VQ56 U4UMC4 A0A0P6IWW3 A0A0P4WPZ7 A0A1J1ILL8 N6U0L5 A0A165AE12 A0A0P6FHE8 A0A0P4WBD5 A0A2P6KTE2 E9GCI3
Pubmed
26354079
22118469
19121390
26227816
17994087
1692392
+ More
10731132 12537572 12537569 1483394 9367444 11923418 14990474 18327897 17550304 12537568 12537573 12537574 16110336 17569856 17569867 22936249 25315136 20920257 15632085 26108605 9461417 17510324 25348373 24495485 20966253 12364791 24438588 25244985 24845553 26823975 25401762 18362917 19820115 20566863 23537049 21292972
10731132 12537572 12537569 1483394 9367444 11923418 14990474 18327897 17550304 12537568 12537573 12537574 16110336 17569856 17569867 22936249 25315136 20920257 15632085 26108605 9461417 17510324 25348373 24495485 20966253 12364791 24438588 25244985 24845553 26823975 25401762 18362917 19820115 20566863 23537049 21292972
EMBL
ODYU01002000
SOQ38959.1
KQ459597
KPI95122.1
KQ460968
KPJ10237.1
+ More
NWSH01000041 PCG80347.1 AGBW02014213 OWR42105.1 BABH01027418 JTDY01001796 KOB72857.1 CM000361 EDX04437.1 X53275 AE014134 AY051981 CM000157 EDW88724.2 AGB92848.1 CH954177 EDV58444.2 CM002910 KMY89381.1 JXJN01015406 JXJN01015407 OUUW01000010 SPP85627.1 AJVK01003811 AJVK01003812 CCAG010007822 JN685590 AFP49901.1 CH479180 EDW28435.1 DS232578 EDS43622.1 ADMH02002211 ETN57778.1 CH379060 EAL33383.3 CH940654 EDW57826.2 CH480818 EDW52340.1 CH916368 EDW03125.1 JRES01000418 KNC31445.1 AH006466 AF023135 AF023136 AAC38845.1 CH933807 EDW12251.2 CH902620 EDV32245.2 CP012523 ALC39135.1 CH477314 EAT43849.1 GAKP01011518 JAC47434.1 ALC38990.1 AXCM01002339 GDHF01021685 JAI30629.1 GAMC01000473 JAC06083.1 AAAB01008944 EAA10082.4 ATLV01014618 KE524975 KFB39249.1 APCN01000643 KU055612 ANC68080.1 CH963857 EDW76675.2 AXCN02000636 LJIG01022862 KRT78338.1 UFQT01000438 SSX24307.1 AXCM01004643 KT981952 AMZ04828.1 KK852854 KDR15027.1 GDHC01012768 JAQ05861.1 GBHO01025345 JAG18259.1 GBHO01025344 JAG18260.1 GDHC01012060 JAQ06569.1 EAA10273.4 ATLV01014619 KFB39250.1 AXCN02000637 KQ971307 EFA11295.1 DS235886 EEB20483.1 MF467212 AXK92489.1 AB986245 BAQ25604.1 KB632384 ERL94267.1 GDIQ01003277 JAN91460.1 GDRN01072352 JAI63565.1 CVRI01000054 CRL00460.1 APGK01047106 APGK01047107 KB741077 ENN74136.1 LRGB01000626 KZS17512.1 GDIQ01050144 JAN44593.1 GDRN01072349 JAI63567.1 MWRG01005575 PRD29568.1 GL732539 EFX82861.1
NWSH01000041 PCG80347.1 AGBW02014213 OWR42105.1 BABH01027418 JTDY01001796 KOB72857.1 CM000361 EDX04437.1 X53275 AE014134 AY051981 CM000157 EDW88724.2 AGB92848.1 CH954177 EDV58444.2 CM002910 KMY89381.1 JXJN01015406 JXJN01015407 OUUW01000010 SPP85627.1 AJVK01003811 AJVK01003812 CCAG010007822 JN685590 AFP49901.1 CH479180 EDW28435.1 DS232578 EDS43622.1 ADMH02002211 ETN57778.1 CH379060 EAL33383.3 CH940654 EDW57826.2 CH480818 EDW52340.1 CH916368 EDW03125.1 JRES01000418 KNC31445.1 AH006466 AF023135 AF023136 AAC38845.1 CH933807 EDW12251.2 CH902620 EDV32245.2 CP012523 ALC39135.1 CH477314 EAT43849.1 GAKP01011518 JAC47434.1 ALC38990.1 AXCM01002339 GDHF01021685 JAI30629.1 GAMC01000473 JAC06083.1 AAAB01008944 EAA10082.4 ATLV01014618 KE524975 KFB39249.1 APCN01000643 KU055612 ANC68080.1 CH963857 EDW76675.2 AXCN02000636 LJIG01022862 KRT78338.1 UFQT01000438 SSX24307.1 AXCM01004643 KT981952 AMZ04828.1 KK852854 KDR15027.1 GDHC01012768 JAQ05861.1 GBHO01025345 JAG18259.1 GBHO01025344 JAG18260.1 GDHC01012060 JAQ06569.1 EAA10273.4 ATLV01014619 KFB39250.1 AXCN02000637 KQ971307 EFA11295.1 DS235886 EEB20483.1 MF467212 AXK92489.1 AB986245 BAQ25604.1 KB632384 ERL94267.1 GDIQ01003277 JAN91460.1 GDRN01072352 JAI63565.1 CVRI01000054 CRL00460.1 APGK01047106 APGK01047107 KB741077 ENN74136.1 LRGB01000626 KZS17512.1 GDIQ01050144 JAN44593.1 GDRN01072349 JAI63567.1 MWRG01005575 PRD29568.1 GL732539 EFX82861.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000005204
UP000037510
+ More
UP000192221 UP000000304 UP000000803 UP000002282 UP000008711 UP000092460 UP000092443 UP000268350 UP000095300 UP000095301 UP000092462 UP000092444 UP000008744 UP000002320 UP000000673 UP000001819 UP000008792 UP000091820 UP000001292 UP000001070 UP000037069 UP000009192 UP000007801 UP000092553 UP000008820 UP000069272 UP000075881 UP000075883 UP000075901 UP000075902 UP000075882 UP000007062 UP000030765 UP000075880 UP000075884 UP000075920 UP000075840 UP000075900 UP000075885 UP000007798 UP000076408 UP000075886 UP000075903 UP000076407 UP000027135 UP000007266 UP000009046 UP000030742 UP000183832 UP000019118 UP000076858 UP000000305
UP000192221 UP000000304 UP000000803 UP000002282 UP000008711 UP000092460 UP000092443 UP000268350 UP000095300 UP000095301 UP000092462 UP000092444 UP000008744 UP000002320 UP000000673 UP000001819 UP000008792 UP000091820 UP000001292 UP000001070 UP000037069 UP000009192 UP000007801 UP000092553 UP000008820 UP000069272 UP000075881 UP000075883 UP000075901 UP000075902 UP000075882 UP000007062 UP000030765 UP000075880 UP000075884 UP000075920 UP000075840 UP000075900 UP000075885 UP000007798 UP000076408 UP000075886 UP000075903 UP000076407 UP000027135 UP000007266 UP000009046 UP000030742 UP000183832 UP000019118 UP000076858 UP000000305
Pfam
PF00230 MIP
Interpro
SUPFAM
SSF81338
SSF81338
Gene 3D
CDD
ProteinModelPortal
A0A2H1VDQ3
A0A194PP89
A0A0N1IEB0
A0A2A4K7Y4
A0A212EKV3
H9J8H5
+ More
A0A0L7LBI9 A0A1W4USY7 B4Q8B5 P23645 B4NYR0 M9PB86 B3N909 A0A0J9QZM1 A0A1B0BJI5 A0A1A9YA33 A0A3B0JU36 A0A1I8Q664 A0A1I8M1G5 A0A1B0D6Y8 L7P705 B4G7T7 B0XAC5 W5J377 Q29NF9 B4M9F0 A0A1A9WW00 B4HWE5 B4JCH1 A0A0L0CGT1 O17044 B4KH40 B3MPC6 A0A0M4EC41 Q17BX1 A0A1I8JSJ9 A0A182JXF3 A0A034VYC6 A0A0M4E804 A0A2K6VB93 A0A182M7A1 A0A0K8UVA6 W8C2H2 A0A182S9X3 A0A182TUN2 A0A182KWM9 Q7Q873 A0A084VMQ5 A0A240PMK8 A0A3F2YW06 A0A1Y9IVN1 A0A1Y9GKD2 A0A1I8JU95 A0A168SE93 A0A1S4GZW8 A0A240PPM5 B4MX36 A0A182Y8J3 A0A182QNB7 A0A0T6ATC6 A0A182VKX8 A0A336M7F0 A0A1Y9J072 A0A182X4Z5 A0A182Y8J1 A0A182MBR0 A0A182IT42 A0A182UCW2 A0A165Y505 A0A067R979 A0A146LC65 A0A182NZ12 A0A0A9XE43 A0A0A9XMC6 A0A146LG91 Q7Q872 A0A084VMQ6 A0A182FZQ0 A0A182PYX1 A0A182II08 A0A1S4GZY8 A0A182VC62 A0A182WQK0 A0A182KWN1 A0A182S4K2 A0A182R1E8 D6W7H3 E0W4C7 A0A346A6C0 A0A0B6VQ56 U4UMC4 A0A0P6IWW3 A0A0P4WPZ7 A0A1J1ILL8 N6U0L5 A0A165AE12 A0A0P6FHE8 A0A0P4WBD5 A0A2P6KTE2 E9GCI3
A0A0L7LBI9 A0A1W4USY7 B4Q8B5 P23645 B4NYR0 M9PB86 B3N909 A0A0J9QZM1 A0A1B0BJI5 A0A1A9YA33 A0A3B0JU36 A0A1I8Q664 A0A1I8M1G5 A0A1B0D6Y8 L7P705 B4G7T7 B0XAC5 W5J377 Q29NF9 B4M9F0 A0A1A9WW00 B4HWE5 B4JCH1 A0A0L0CGT1 O17044 B4KH40 B3MPC6 A0A0M4EC41 Q17BX1 A0A1I8JSJ9 A0A182JXF3 A0A034VYC6 A0A0M4E804 A0A2K6VB93 A0A182M7A1 A0A0K8UVA6 W8C2H2 A0A182S9X3 A0A182TUN2 A0A182KWM9 Q7Q873 A0A084VMQ5 A0A240PMK8 A0A3F2YW06 A0A1Y9IVN1 A0A1Y9GKD2 A0A1I8JU95 A0A168SE93 A0A1S4GZW8 A0A240PPM5 B4MX36 A0A182Y8J3 A0A182QNB7 A0A0T6ATC6 A0A182VKX8 A0A336M7F0 A0A1Y9J072 A0A182X4Z5 A0A182Y8J1 A0A182MBR0 A0A182IT42 A0A182UCW2 A0A165Y505 A0A067R979 A0A146LC65 A0A182NZ12 A0A0A9XE43 A0A0A9XMC6 A0A146LG91 Q7Q872 A0A084VMQ6 A0A182FZQ0 A0A182PYX1 A0A182II08 A0A1S4GZY8 A0A182VC62 A0A182WQK0 A0A182KWN1 A0A182S4K2 A0A182R1E8 D6W7H3 E0W4C7 A0A346A6C0 A0A0B6VQ56 U4UMC4 A0A0P6IWW3 A0A0P4WPZ7 A0A1J1ILL8 N6U0L5 A0A165AE12 A0A0P6FHE8 A0A0P4WBD5 A0A2P6KTE2 E9GCI3
PDB
3CN6
E-value=8.15491e-09,
Score=138
Ontologies
GO
PANTHER
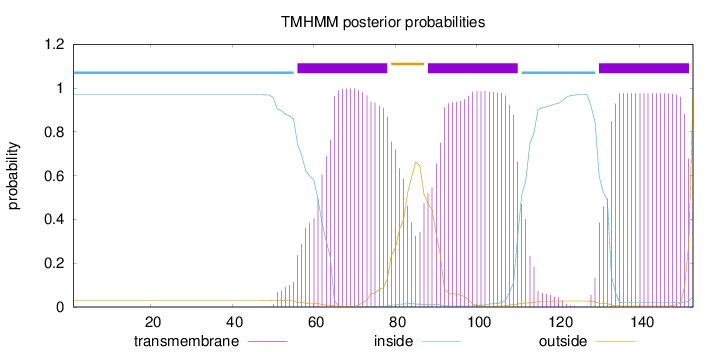
Topology
Subcellular location
Membrane
Length:
153
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.28954
Exp number, first 60 AAs:
2.13173
Total prob of N-in:
0.96949
inside
1 - 55
TMhelix
56 - 78
outside
79 - 87
TMhelix
88 - 110
inside
111 - 129
TMhelix
130 - 152
outside
153 - 153
Population Genetic Test Statistics
Pi
281.102976
Theta
163.493554
Tajima's D
2.450601
CLR
0.43538
CSRT
0.941402929853507
Interpretation
Uncertain
