Gene
KWMTBOMO03071 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005755
Annotation
small_heat_shock_protein_[Spodoptera_litura]
Location in the cell
Extracellular Reliability : 1.537
Sequence
CDS
ATGTTCTCACCGCGTCTCTTTGCGGTTTTCATCGCTTTCGCCGGTTTCGCAACGGTAACGGCTTTGCCTGCCCGGGACAAAGAAGTTATCGCGCAGCCCGTCTCCATTACAGATGACCAGTTCACGTTCCCATGGCTGAACCTTTCGCCATTCAGTCTGTTTGCTCCTTTGTGGAAGCTGATCCCCACTTTTGTGGACATCGGGCCAAGAATCTACGCTGACGATGACAAGTTCAAGGTCGTTGTCAACGTTAAGGACTACAAGAAAGACAACCTTAAAGTTAAAGTAAAGGGAGACTTTATCTTCGTCCAAGGATCGCAAGAAGCTAAAGAAGACGACCATGACGTGTTCGCCAGCCAGTTCTTCCACACCTATAGCCTACCTGTGAACTCGAGTGCTGCTGATGTTACGGCTGAGCTGACTTCCGATGGATACCTCGTCGTGACGGCGCCGATCAGCGAAAATGTTGATAAAACGAAAAATACAGAGCGCGTTGTACCTATTGTCGAGACTGGCGCTCCGTACAAGAAGGACGAGCCTGTAGAAAAGACGACAGTAGAAACCTTGGACGTTTCTACGACTCAGGAGCCGAAGACATCAGCTCCAGCGGTAGTGACTGCAGCACCGGAACCCGAGGAGAAGAAAGAACCGACCACGCCCTCCGAACTCGATGAAGCGACAGAAAAAGACAACGTGATCCCACACGGAAACGAAGTTTCTGCTTAA
Protein
MFSPRLFAVFIAFAGFATVTALPARDKEVIAQPVSITDDQFTFPWLNLSPFSLFAPLWKLIPTFVDIGPRIYADDDKFKVVVNVKDYKKDNLKVKVKGDFIFVQGSQEAKEDDHDVFASQFFHTYSLPVNSSAADVTAELTSDGYLVVTAPISENVDKTKNTERVVPIVETGAPYKKDEPVEKTTVETLDVSTTQEPKTSAPAVVTAAPEPEEKKEPTTPSELDEATEKDNVIPHGNEVSA
Summary
Similarity
Belongs to the small heat shock protein (HSP20) family.
Uniprot
EMBL
BABH01027461
ODYU01001230
SOQ37158.1
HM046616
ADK55523.1
KC689792
+ More
KX845560 KZ150087 AGQ42770.1 ATB54991.1 PZC73752.1 RBVL01000268 RKO09897.1 MG992393 AXY94831.1 KP843899 AKS40078.1 KX958481 ASQ43197.1 JTDY01003326 KOB69737.1 KQ459597 KPI95192.1 KQ460970 KPJ10164.1 AGBW02008445 OWR53388.1 KJ461925 AHW45927.1 AGBW02011158 OWR47132.1 KX958480 ASQ43196.1
KX845560 KZ150087 AGQ42770.1 ATB54991.1 PZC73752.1 RBVL01000268 RKO09897.1 MG992393 AXY94831.1 KP843899 AKS40078.1 KX958481 ASQ43197.1 JTDY01003326 KOB69737.1 KQ459597 KPI95192.1 KQ460970 KPJ10164.1 AGBW02008445 OWR53388.1 KJ461925 AHW45927.1 AGBW02011158 OWR47132.1 KX958480 ASQ43196.1
Proteomes
PRIDE
Pfam
PF00011 HSP20
Interpro
SUPFAM
SSF49764
SSF49764
Gene 3D
ProteinModelPortal
PDB
3N3E
E-value=3.31239e-05,
Score=110
Ontologies
PATHWAY
GO
Topology
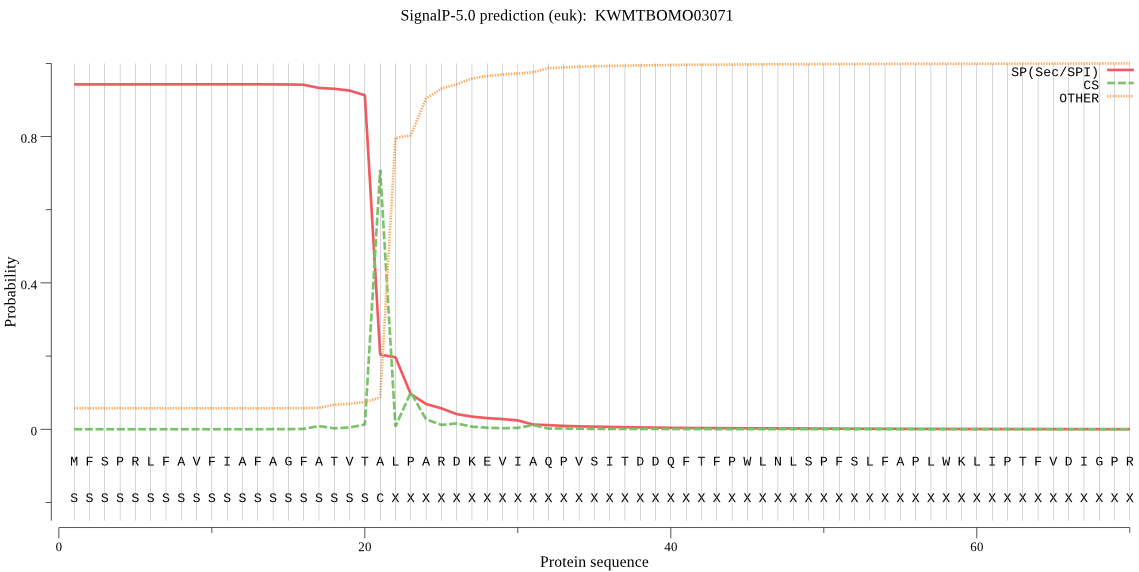
SignalP
Position: 1 - 21,
Likelihood: 0.942257
Length:
241
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.90936
Exp number, first 60 AAs:
12.50292
Total prob of N-in:
0.50467
POSSIBLE N-term signal
sequence
outside
1 - 241

Population Genetic Test Statistics
Pi
243.321146
Theta
215.58176
Tajima's D
0.405132
CLR
160.257536
CSRT
0.484225788710564
Interpretation
Uncertain
