Gene
KWMTBOMO03064
Pre Gene Modal
BGIBMGA005750
Annotation
PREDICTED:_sulfite_oxidase_isoform_X1_[Bombyx_mori]
Full name
Probable sulfite oxidase, mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.87 Mitochondrial Reliability : 1.519
Sequence
CDS
ATGTCTTTAAAAAAAATACTACTGGGCACTGTATTCCTAAGAAACAGATACATAGGATTATCTTCAGTTTCAACAATAACTGACAAAAAATACAGATTTGATGAGAGTGAAGGTTTTTTATTTAGAACCAAAGGAAAGATTCTTACAGCAGCTGTCTTCAGTTCTGTGCTTATTGGAGCTGCTGTTAATGAACAGATAAAAGATAAAAATGTCAAATCACAATCTAATGATGTTGTTATTGAAGCTGGAGAGAAATTACCTGATCTTCCAACATATACAGCAGAAGAAGTCAGCAAACACGGAGACAAAAATAGTTTTTGGGTGACATACAAACATGGTGTTTATGATGTAACAAAATTCCTTCCGTCCCATCCTGGAGGGGAACAGATCATGAACGCAGGGGGACTCAGTATAGAGCCATTTTGGAATGTGTATGGGATGCATAAAACTCCAGAGATTTATAAGTTACTTGAATCGTACAGGATTGGCAACCTCCATGACGACGATGTAATAGATCACTCAGATGATGAATTGTGGGTCAAAGAGCCAGTTAGGGATAACAGGCTCATAGTTAAAACCGCCAAGCCATTCAATGCTGAAATACCTGCAAAATATCAAATTAAATTCTTCGATACTCCCAACGAGTTATTTTACGTGCGCCAACACATGCCTGTTCCAGAGCTGGACGCGGACGATCACCGTTTGAAGGTCACCGTCAAGGGATCTAAGATTCGTACCGAAGAGTTCAGTCTCCGCGACTTGGATAAGTTCCGGAGGGTTACCACCAGGGCGGCTCTGATGTGTGCCGGGAATCGACGCAGTGAGATGAATGAGGTGAAACCGGTGAAAGGTATCGGCTGGATGGGCGGGGCGATCAGCAACGCGGTGTGGGGCGGAGTGTATCTCAGAGACGTCCTCTTGTACAGCGGCGTGGACCCGGAAGACACTAAAGGGAAACACGTTATTTTCACAGGCGCAGACATAGACGCGACCGGCGTTAATTTCAGCACGTCGATACCGCTGGACATGGCGATGAACCCCAGCAGCGGCATCCTGCTCGCCACCACCATGAACGGCCACCCCCTGCCCCCCGACCACGGCCGCCCCCTGCGCGTGGTGGTCCCCGGGGCGCCCGCCGTGCGGAGCGTCAAGTGGCTGCAATCCATCACCGTCTCCGAAGACGAGAGTCCCTCGCACTGGCACCAGAAAGACTACAGAGCCTTCAACCCGTCCAAGACGTGGGAGACGGCGGACTTCGCCACCGCGCCGCCCGTGTACAGCCTGCCCGTCACCTCCGCGATCTGCGACCCCGCCGACGGTGACGTTATCAGTGTCAAGAACGGAGCTGTTCAAGTTCGAGGCTACGCGTACTCCGGCGGCGGGGCGAAGATCGTCCGCGTGGACGTGTCGGCGGACGGCGGGCGCAGCTGGTGGGAGGCGCGCGAGCTGCAGGCCGACGCCGCGCCGCCGCAGCAGCACTACGGCTGGACCCTGTGGACGGCCAGCGTGCCCGTCGCGACACAACAGAAGGAGGTGGAACTATGGGCTAAAGCTACTGACAGCAATTTCAATACGCAGCCAGAACGATTCAACGACATTTGGAACATTAGAGGAATACTCAGCAACGCCTACCACAGAATCAAAATTAAACTTGTACATTAA
Protein
MSLKKILLGTVFLRNRYIGLSSVSTITDKKYRFDESEGFLFRTKGKILTAAVFSSVLIGAAVNEQIKDKNVKSQSNDVVIEAGEKLPDLPTYTAEEVSKHGDKNSFWVTYKHGVYDVTKFLPSHPGGEQIMNAGGLSIEPFWNVYGMHKTPEIYKLLESYRIGNLHDDDVIDHSDDELWVKEPVRDNRLIVKTAKPFNAEIPAKYQIKFFDTPNELFYVRQHMPVPELDADDHRLKVTVKGSKIRTEEFSLRDLDKFRRVTTRAALMCAGNRRSEMNEVKPVKGIGWMGGAISNAVWGGVYLRDVLLYSGVDPEDTKGKHVIFTGADIDATGVNFSTSIPLDMAMNPSSGILLATTMNGHPLPPDHGRPLRVVVPGAPAVRSVKWLQSITVSEDESPSHWHQKDYRAFNPSKTWETADFATAPPVYSLPVTSAICDPADGDVISVKNGAVQVRGYAYSGGGAKIVRVDVSADGGRSWWEARELQADAAPPQQHYGWTLWTASVPVATQQKEVELWAKATDSNFNTQPERFNDIWNIRGILSNAYHRIKIKLVH
Summary
Catalytic Activity
H2O + O2 + sulfite = H2O2 + sulfate
Cofactor
Mo-molybdopterin
heme b
heme b
Subunit
Homodimer.
Similarity
Belongs to the cytochrome b5 family.
Keywords
Complete proteome
Heme
Iron
Metal-binding
Mitochondrion
Molybdenum
Oxidoreductase
Reference proteome
Transit peptide
Feature
chain Probable sulfite oxidase, mitochondrial
Uniprot
A0A2W1BPZ2
A0A2A4JWW6
A0A2H1VZR0
A0A2A4JVX8
A0A194PP44
A0A0N1I7H6
+ More
A0A212F7P2 A0A1B6MLX5 D6WC52 A0A2M4CYI5 A0A2J7QY53 A0A1B6CB46 A0A1B6DI64 A0A2M4BHI4 W5JMG7 A0A182QBC5 A0A182FKC0 A0A2M4APZ7 A0A2M4AP70 A0A2M4APT3 A0A2M4BM17 W8C9R9 A0A087T7K7 A0A0M8ZWU8 A0A1L8E0I5 E9IVG1 B0WBF7 A0A0L0BV58 B4M2Z6 B4L5I4 A0A3L8D4N3 A0A026VVB7 A0A195C674 A0A0A1XP73 E2B6H6 A0A1Q3F5Y9 A0A2P2I5P2 A0A0J7KFT1 A0A1L8EIL4 A0A084VSJ5 A0A182SMK8 A0A182NC90 A0A1I8MUV9 A0A182PJB2 A0A2P8YMI9 A0A195FUJ1 E2AF16 D3TPZ1 A0A1B6BW31 A0A1B0G5H6 B4JX09 A0A1S4G1D6 Q16H84 A0A1I8P527 A0A182YKM0 A0A182RYX0 A0A1B0CUW3 A0A158N9U2 A0A182WFX0 A0A1A9ULH6 Q7QGS7 A0A1A9X7Q6 A0A182KVL5 A0A1B0AIW0 A0A182MKG9 A0A0V0G9T2 A0A195BBJ9 A0A182TCY0 A0A182VI56 A0A023F6G2 A0A182X410 T1I4V4 F4WLY3 A0A3B0JZU4 A0A182I5H9 A0A1W4U9X0 A0A034WR20 A0A182IKS0 A0A1B0AX09 A0A1A9W238 A0A1B6GR39 A0A0M3QZN5 A0A336LNS1 A0A088ADD5 A0A195DCQ6 K7J789 A0A310SF24 Q9VWP4 A0A0M5J633 Q29GE9 Q16GU7 A0A2A3EEW8 T1JZC5 A0A182H3F0 A0A2L2Y2R1 U4U2I2 B4PWG2 A0A224Z0B4 E9G6E0
A0A212F7P2 A0A1B6MLX5 D6WC52 A0A2M4CYI5 A0A2J7QY53 A0A1B6CB46 A0A1B6DI64 A0A2M4BHI4 W5JMG7 A0A182QBC5 A0A182FKC0 A0A2M4APZ7 A0A2M4AP70 A0A2M4APT3 A0A2M4BM17 W8C9R9 A0A087T7K7 A0A0M8ZWU8 A0A1L8E0I5 E9IVG1 B0WBF7 A0A0L0BV58 B4M2Z6 B4L5I4 A0A3L8D4N3 A0A026VVB7 A0A195C674 A0A0A1XP73 E2B6H6 A0A1Q3F5Y9 A0A2P2I5P2 A0A0J7KFT1 A0A1L8EIL4 A0A084VSJ5 A0A182SMK8 A0A182NC90 A0A1I8MUV9 A0A182PJB2 A0A2P8YMI9 A0A195FUJ1 E2AF16 D3TPZ1 A0A1B6BW31 A0A1B0G5H6 B4JX09 A0A1S4G1D6 Q16H84 A0A1I8P527 A0A182YKM0 A0A182RYX0 A0A1B0CUW3 A0A158N9U2 A0A182WFX0 A0A1A9ULH6 Q7QGS7 A0A1A9X7Q6 A0A182KVL5 A0A1B0AIW0 A0A182MKG9 A0A0V0G9T2 A0A195BBJ9 A0A182TCY0 A0A182VI56 A0A023F6G2 A0A182X410 T1I4V4 F4WLY3 A0A3B0JZU4 A0A182I5H9 A0A1W4U9X0 A0A034WR20 A0A182IKS0 A0A1B0AX09 A0A1A9W238 A0A1B6GR39 A0A0M3QZN5 A0A336LNS1 A0A088ADD5 A0A195DCQ6 K7J789 A0A310SF24 Q9VWP4 A0A0M5J633 Q29GE9 Q16GU7 A0A2A3EEW8 T1JZC5 A0A182H3F0 A0A2L2Y2R1 U4U2I2 B4PWG2 A0A224Z0B4 E9G6E0
EC Number
1.8.3.1
Pubmed
28756777
26354079
22118469
18362917
19820115
20920257
+ More
23761445 24495485 21282665 26108605 17994087 30249741 24508170 25830018 20798317 24438588 25315136 29403074 20353571 17510324 25244985 21347285 12364791 20966253 25474469 21719571 25348373 20075255 10731132 12537572 12537569 15632085 26483478 26561354 23537049 17550304 28797301 21292972
23761445 24495485 21282665 26108605 17994087 30249741 24508170 25830018 20798317 24438588 25315136 29403074 20353571 17510324 25244985 21347285 12364791 20966253 25474469 21719571 25348373 20075255 10731132 12537572 12537569 15632085 26483478 26561354 23537049 17550304 28797301 21292972
EMBL
KZ150087
PZC73763.1
NWSH01000470
PCG76198.1
ODYU01005322
SOQ46052.1
+ More
PCG76197.1 KQ459597 KPI95206.1 KQ460970 KPJ10152.1 AGBW02009848 OWR49752.1 GEBQ01019539 GEBQ01018339 GEBQ01003029 JAT20438.1 JAT21638.1 JAT36948.1 KQ971316 EEZ97850.1 GGFL01006215 MBW70393.1 NEVH01009368 PNF33513.1 GEDC01026783 JAS10515.1 GEDC01026297 GEDC01011973 JAS11001.1 JAS25325.1 GGFJ01003312 MBW52453.1 ADMH02001144 ETN63944.1 AXCN02001762 GGFK01009367 MBW42688.1 GGFK01009248 MBW42569.1 GGFK01009297 MBW42618.1 GGFJ01004988 MBW54129.1 GAMC01006406 GAMC01006405 JAC00150.1 KK113815 KFM61096.1 KQ435812 KOX72740.1 GFDF01001900 JAV12184.1 GL766234 EFZ15438.1 DS231879 EDS42396.1 JRES01001291 KNC23906.1 CH940651 EDW65171.1 CH933811 EDW06443.1 QOIP01000013 RLU15184.1 KK107796 EZA47687.1 KQ978220 KYM96364.1 GBXI01001550 JAD12742.1 GL445973 EFN88720.1 GFDL01012061 JAV22984.1 IACF01003743 LAB69353.1 LBMM01008146 KMQ89109.1 GFDG01000340 JAV18459.1 ATLV01016042 ATLV01016043 KE525054 KFB40939.1 PYGN01000491 PSN45459.1 KQ981264 KYN43962.1 GL438984 EFN67971.1 EZ423493 ADD19769.1 GEDC01031791 JAS05507.1 CCAG010002752 CH916376 EDV95285.1 CH478192 EAT33616.1 AJWK01029714 AJWK01029715 ADTU01001413 AAAB01008823 EAA05527.4 AXCM01005795 GECL01001981 JAP04143.1 KQ976528 KYM81913.1 GBBI01001880 JAC16832.1 ACPB03006148 GL888217 EGI64627.1 OUUW01000011 SPP86593.1 APCN01003445 GAKP01001863 JAC57089.1 JXJN01004967 GECZ01004868 JAS64901.1 CP012528 ALC49689.1 UFQT01000094 SSX19656.1 KQ980989 KYN10663.1 KQ760549 OAD59884.1 AE014298 AY069352 ALC48968.1 CH379064 EAL32160.2 CH478233 EAT33474.1 KZ288271 PBC29822.1 CAEY01001120 JXUM01107365 JXUM01107366 KQ565135 KXJ71297.1 IAAA01009366 LAA02441.1 KB631625 ERL84806.1 CM000162 EDX02780.1 GFPF01012131 MAA23277.1 GL732533 EFX84919.1
PCG76197.1 KQ459597 KPI95206.1 KQ460970 KPJ10152.1 AGBW02009848 OWR49752.1 GEBQ01019539 GEBQ01018339 GEBQ01003029 JAT20438.1 JAT21638.1 JAT36948.1 KQ971316 EEZ97850.1 GGFL01006215 MBW70393.1 NEVH01009368 PNF33513.1 GEDC01026783 JAS10515.1 GEDC01026297 GEDC01011973 JAS11001.1 JAS25325.1 GGFJ01003312 MBW52453.1 ADMH02001144 ETN63944.1 AXCN02001762 GGFK01009367 MBW42688.1 GGFK01009248 MBW42569.1 GGFK01009297 MBW42618.1 GGFJ01004988 MBW54129.1 GAMC01006406 GAMC01006405 JAC00150.1 KK113815 KFM61096.1 KQ435812 KOX72740.1 GFDF01001900 JAV12184.1 GL766234 EFZ15438.1 DS231879 EDS42396.1 JRES01001291 KNC23906.1 CH940651 EDW65171.1 CH933811 EDW06443.1 QOIP01000013 RLU15184.1 KK107796 EZA47687.1 KQ978220 KYM96364.1 GBXI01001550 JAD12742.1 GL445973 EFN88720.1 GFDL01012061 JAV22984.1 IACF01003743 LAB69353.1 LBMM01008146 KMQ89109.1 GFDG01000340 JAV18459.1 ATLV01016042 ATLV01016043 KE525054 KFB40939.1 PYGN01000491 PSN45459.1 KQ981264 KYN43962.1 GL438984 EFN67971.1 EZ423493 ADD19769.1 GEDC01031791 JAS05507.1 CCAG010002752 CH916376 EDV95285.1 CH478192 EAT33616.1 AJWK01029714 AJWK01029715 ADTU01001413 AAAB01008823 EAA05527.4 AXCM01005795 GECL01001981 JAP04143.1 KQ976528 KYM81913.1 GBBI01001880 JAC16832.1 ACPB03006148 GL888217 EGI64627.1 OUUW01000011 SPP86593.1 APCN01003445 GAKP01001863 JAC57089.1 JXJN01004967 GECZ01004868 JAS64901.1 CP012528 ALC49689.1 UFQT01000094 SSX19656.1 KQ980989 KYN10663.1 KQ760549 OAD59884.1 AE014298 AY069352 ALC48968.1 CH379064 EAL32160.2 CH478233 EAT33474.1 KZ288271 PBC29822.1 CAEY01001120 JXUM01107365 JXUM01107366 KQ565135 KXJ71297.1 IAAA01009366 LAA02441.1 KB631625 ERL84806.1 CM000162 EDX02780.1 GFPF01012131 MAA23277.1 GL732533 EFX84919.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000007266
UP000235965
+ More
UP000000673 UP000075886 UP000069272 UP000054359 UP000053105 UP000002320 UP000037069 UP000008792 UP000009192 UP000279307 UP000053097 UP000078542 UP000008237 UP000036403 UP000030765 UP000075901 UP000075884 UP000095301 UP000075885 UP000245037 UP000078541 UP000000311 UP000092444 UP000001070 UP000008820 UP000095300 UP000076408 UP000075900 UP000092461 UP000005205 UP000075920 UP000078200 UP000007062 UP000092443 UP000075882 UP000092445 UP000075883 UP000078540 UP000075902 UP000075903 UP000076407 UP000015103 UP000007755 UP000268350 UP000075840 UP000192221 UP000075880 UP000092460 UP000091820 UP000092553 UP000005203 UP000078492 UP000002358 UP000000803 UP000001819 UP000242457 UP000015104 UP000069940 UP000249989 UP000030742 UP000002282 UP000000305
UP000000673 UP000075886 UP000069272 UP000054359 UP000053105 UP000002320 UP000037069 UP000008792 UP000009192 UP000279307 UP000053097 UP000078542 UP000008237 UP000036403 UP000030765 UP000075901 UP000075884 UP000095301 UP000075885 UP000245037 UP000078541 UP000000311 UP000092444 UP000001070 UP000008820 UP000095300 UP000076408 UP000075900 UP000092461 UP000005205 UP000075920 UP000078200 UP000007062 UP000092443 UP000075882 UP000092445 UP000075883 UP000078540 UP000075902 UP000075903 UP000076407 UP000015103 UP000007755 UP000268350 UP000075840 UP000192221 UP000075880 UP000092460 UP000091820 UP000092553 UP000005203 UP000078492 UP000002358 UP000000803 UP000001819 UP000242457 UP000015104 UP000069940 UP000249989 UP000030742 UP000002282 UP000000305
PRIDE
Interpro
IPR000572
OxRdtase_Mopterin-bd_dom
+ More
IPR036374 OxRdtase_Mopterin-bd_sf
IPR018506 Cyt_B5_heme-BS
IPR005066 MoCF_OxRdtse_dimer
IPR001199 Cyt_B5-like_heme/steroid-bd
IPR014756 Ig_E-set
IPR036400 Cyt_B5-like_heme/steroid_sf
IPR008335 Mopterin_OxRdtase_euk
IPR022407 OxRdtase_Mopterin_BS
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
IPR036374 OxRdtase_Mopterin-bd_sf
IPR018506 Cyt_B5_heme-BS
IPR005066 MoCF_OxRdtse_dimer
IPR001199 Cyt_B5-like_heme/steroid-bd
IPR014756 Ig_E-set
IPR036400 Cyt_B5-like_heme/steroid_sf
IPR008335 Mopterin_OxRdtase_euk
IPR022407 OxRdtase_Mopterin_BS
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000719 Prot_kinase_dom
Gene 3D
ProteinModelPortal
A0A2W1BPZ2
A0A2A4JWW6
A0A2H1VZR0
A0A2A4JVX8
A0A194PP44
A0A0N1I7H6
+ More
A0A212F7P2 A0A1B6MLX5 D6WC52 A0A2M4CYI5 A0A2J7QY53 A0A1B6CB46 A0A1B6DI64 A0A2M4BHI4 W5JMG7 A0A182QBC5 A0A182FKC0 A0A2M4APZ7 A0A2M4AP70 A0A2M4APT3 A0A2M4BM17 W8C9R9 A0A087T7K7 A0A0M8ZWU8 A0A1L8E0I5 E9IVG1 B0WBF7 A0A0L0BV58 B4M2Z6 B4L5I4 A0A3L8D4N3 A0A026VVB7 A0A195C674 A0A0A1XP73 E2B6H6 A0A1Q3F5Y9 A0A2P2I5P2 A0A0J7KFT1 A0A1L8EIL4 A0A084VSJ5 A0A182SMK8 A0A182NC90 A0A1I8MUV9 A0A182PJB2 A0A2P8YMI9 A0A195FUJ1 E2AF16 D3TPZ1 A0A1B6BW31 A0A1B0G5H6 B4JX09 A0A1S4G1D6 Q16H84 A0A1I8P527 A0A182YKM0 A0A182RYX0 A0A1B0CUW3 A0A158N9U2 A0A182WFX0 A0A1A9ULH6 Q7QGS7 A0A1A9X7Q6 A0A182KVL5 A0A1B0AIW0 A0A182MKG9 A0A0V0G9T2 A0A195BBJ9 A0A182TCY0 A0A182VI56 A0A023F6G2 A0A182X410 T1I4V4 F4WLY3 A0A3B0JZU4 A0A182I5H9 A0A1W4U9X0 A0A034WR20 A0A182IKS0 A0A1B0AX09 A0A1A9W238 A0A1B6GR39 A0A0M3QZN5 A0A336LNS1 A0A088ADD5 A0A195DCQ6 K7J789 A0A310SF24 Q9VWP4 A0A0M5J633 Q29GE9 Q16GU7 A0A2A3EEW8 T1JZC5 A0A182H3F0 A0A2L2Y2R1 U4U2I2 B4PWG2 A0A224Z0B4 E9G6E0
A0A212F7P2 A0A1B6MLX5 D6WC52 A0A2M4CYI5 A0A2J7QY53 A0A1B6CB46 A0A1B6DI64 A0A2M4BHI4 W5JMG7 A0A182QBC5 A0A182FKC0 A0A2M4APZ7 A0A2M4AP70 A0A2M4APT3 A0A2M4BM17 W8C9R9 A0A087T7K7 A0A0M8ZWU8 A0A1L8E0I5 E9IVG1 B0WBF7 A0A0L0BV58 B4M2Z6 B4L5I4 A0A3L8D4N3 A0A026VVB7 A0A195C674 A0A0A1XP73 E2B6H6 A0A1Q3F5Y9 A0A2P2I5P2 A0A0J7KFT1 A0A1L8EIL4 A0A084VSJ5 A0A182SMK8 A0A182NC90 A0A1I8MUV9 A0A182PJB2 A0A2P8YMI9 A0A195FUJ1 E2AF16 D3TPZ1 A0A1B6BW31 A0A1B0G5H6 B4JX09 A0A1S4G1D6 Q16H84 A0A1I8P527 A0A182YKM0 A0A182RYX0 A0A1B0CUW3 A0A158N9U2 A0A182WFX0 A0A1A9ULH6 Q7QGS7 A0A1A9X7Q6 A0A182KVL5 A0A1B0AIW0 A0A182MKG9 A0A0V0G9T2 A0A195BBJ9 A0A182TCY0 A0A182VI56 A0A023F6G2 A0A182X410 T1I4V4 F4WLY3 A0A3B0JZU4 A0A182I5H9 A0A1W4U9X0 A0A034WR20 A0A182IKS0 A0A1B0AX09 A0A1A9W238 A0A1B6GR39 A0A0M3QZN5 A0A336LNS1 A0A088ADD5 A0A195DCQ6 K7J789 A0A310SF24 Q9VWP4 A0A0M5J633 Q29GE9 Q16GU7 A0A2A3EEW8 T1JZC5 A0A182H3F0 A0A2L2Y2R1 U4U2I2 B4PWG2 A0A224Z0B4 E9G6E0
PDB
1SOX
E-value=1.24197e-107,
Score=998
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion intermembrane space
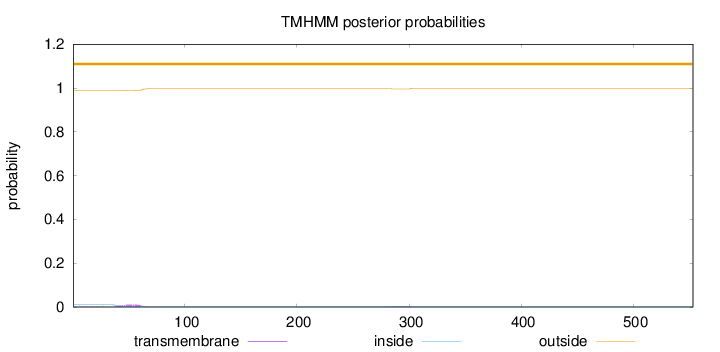
Length:
553
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.277710000000001
Exp number, first 60 AAs:
0.20309
Total prob of N-in:
0.01013
outside
1 - 553
Population Genetic Test Statistics
Pi
174.269629
Theta
142.834205
Tajima's D
0.669925
CLR
0.426804
CSRT
0.565121743912804
Interpretation
Uncertain
