Gene
KWMTBOMO03058
Annotation
PREDICTED:_atrial_natriuretic_peptide-converting_enzyme_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.846
Sequence
CDS
ATGGCAGCCATTGAATTAAATCGGGGAGACTTATGGCTCCTCGTTACAGAATGTGGCAAGCGTCGTACGGTCGGAGGTGGAACAAAGCGCATCGTTGGAGGAGTAGAAGCATCTCCTGGAGACTGGCCCTTTCTGGCCGCGATTCTGGGAGGGCCGGAGGAGGTGTTCTACTGCGCCGGTGTACTCATTGCTGATCAGTGGATACTTACTGCCTCTCATTGCGTTGGCAATCACTCAGACGTGAATGGATGGACCATCCAGCTGGGTATAACTAGACGACGCTCTCATGCCTACTACGGTCAGAAGGTGAAGGTGCGTCGGGTGGTGCCACACCCGCAGTACAACGTTGGTGTCGCTCATGATAACGACATCGCTTTATTCCAGTTGGCGGTGAGGGTGCGCTACCACGAGCAGCTGTCGCCGGTGTGCCTGCCGCCCGCCGACCGGCCGCTGCCCGCCGGCACGCTCTGCACCGTCATCGGGTGGGGGAAACGAGACGATAAGGACATGTCTGAATACGAGCCGGCAGTGAATGAAGTGGAAGTGCCCGTGCTGCATCGCGACCTCTGCAACCAGTGGCTGGAGCATCGGGACCTCAACGTCACCGAGGGGATGATCTGCGCCGGGTACCCGGAGGGAGGAAAAGATGCTTGTCAGGGCGATTCCGGCGGGCCGCTGCTCTGTAAGGATCCCGACGACTCCTCGCGCTGGTACGTCGGCGGCATCGTGTCTTGGGGCATCAAATGCGCGCACCCCCGCCTCCCGGGAGTCTACGCGTACGTCTCCAAATATATACCGTGGATACAGAAACATATTAGTCTTTATAACGACGACAACGCCAAGTCTGAAGACGTTTAG
Protein
MAAIELNRGDLWLLVTECGKRRTVGGGTKRIVGGVEASPGDWPFLAAILGGPEEVFYCAGVLIADQWILTASHCVGNHSDVNGWTIQLGITRRRSHAYYGQKVKVRRVVPHPQYNVGVAHDNDIALFQLAVRVRYHEQLSPVCLPPADRPLPAGTLCTVIGWGKRDDKDMSEYEPAVNEVEVPVLHRDLCNQWLEHRDLNVTEGMICAGYPEGGKDACQGDSGGPLLCKDPDDSSRWYVGGIVSWGIKCAHPRLPGVYAYVSKYIPWIQKHISLYNDDNAKSEDV
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A2H1V872
A0A212F7R4
A0A0N1IE78
A0A194PPH0
A0A2W1BS65
A0A067RJW0
+ More
N6T216 D6W7Q1 A0A195BXT6 A0A2J7PTF4 A0A195CV41 A0A0A9Y6G6 A0A158NQ02 T1I1Y6 A0A1B6GI55 K7IQW1 F4X817 A0A151WZ95 A0A0C9RF23 A0A3L8D8M9 E1ZW18 A0A026WWV7 A0A151JQ49 A0A195ETY3 A0A088ARG2 E0VUT5 A0A182GGD0 Q16SA2 A0A182S7G8 A0A182Q4B6 A0A023F359 A0A068F4F8 A0A182R978 A0A1B0C9Z5 A0A336KNG9 A0A182TY97 Q7Q554 A0A1J1I006 A0A182I4L0 A0A182LKB7 A0A182PDL4 A0A182X4L3 A0A182VBY8 A0A1A9X3N6 A0A182LTZ4 A0A182NI76 A0A182WBN6 A0A1S4ESL9 A0A1I8PR60 A0A1I8PRA4 A0A1I8MUQ7 A0A182J3Y1 A0A1W4VDJ1 A0A0J9R7H2 B3N9X1 A1Z709 B4P1R7 B4HR36 A0A182FEA1 A0A034VA32 B4KPP9 B4N657 Q28YJ3 A0A0R3NNY5 B4H8J4 A0A0M4EVS7 B4LJ88 B3MHB8 B4J7Y2 A0A1B0D2D8 A0A154P8C6 A0A2P8Z1M6 A0A336MC12 J9KVA0 A0A084WMG8 T1KG36 A0A131ZYS7 A0A1Y3BS63 A0A210QQ31 A0A3B4T7E3 A0A3B4T6Y7 A0A3B4T6V1 A0A3B4T6M8 A0A3B4T6L2 A0A2C9KCV1
N6T216 D6W7Q1 A0A195BXT6 A0A2J7PTF4 A0A195CV41 A0A0A9Y6G6 A0A158NQ02 T1I1Y6 A0A1B6GI55 K7IQW1 F4X817 A0A151WZ95 A0A0C9RF23 A0A3L8D8M9 E1ZW18 A0A026WWV7 A0A151JQ49 A0A195ETY3 A0A088ARG2 E0VUT5 A0A182GGD0 Q16SA2 A0A182S7G8 A0A182Q4B6 A0A023F359 A0A068F4F8 A0A182R978 A0A1B0C9Z5 A0A336KNG9 A0A182TY97 Q7Q554 A0A1J1I006 A0A182I4L0 A0A182LKB7 A0A182PDL4 A0A182X4L3 A0A182VBY8 A0A1A9X3N6 A0A182LTZ4 A0A182NI76 A0A182WBN6 A0A1S4ESL9 A0A1I8PR60 A0A1I8PRA4 A0A1I8MUQ7 A0A182J3Y1 A0A1W4VDJ1 A0A0J9R7H2 B3N9X1 A1Z709 B4P1R7 B4HR36 A0A182FEA1 A0A034VA32 B4KPP9 B4N657 Q28YJ3 A0A0R3NNY5 B4H8J4 A0A0M4EVS7 B4LJ88 B3MHB8 B4J7Y2 A0A1B0D2D8 A0A154P8C6 A0A2P8Z1M6 A0A336MC12 J9KVA0 A0A084WMG8 T1KG36 A0A131ZYS7 A0A1Y3BS63 A0A210QQ31 A0A3B4T7E3 A0A3B4T6Y7 A0A3B4T6V1 A0A3B4T6M8 A0A3B4T6L2 A0A2C9KCV1
Pubmed
22118469
26354079
28756777
24845553
23537049
18362917
+ More
19820115 25401762 21347285 20075255 21719571 30249741 20798317 24508170 20566863 26483478 17510324 25474469 24952583 12364791 20966253 25315136 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 15632085 23185243 18057021 29403074 24438588 26555130 28812685 15562597
19820115 25401762 21347285 20075255 21719571 30249741 20798317 24508170 20566863 26483478 17510324 25474469 24952583 12364791 20966253 25315136 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 15632085 23185243 18057021 29403074 24438588 26555130 28812685 15562597
EMBL
ODYU01001183
SOQ37048.1
AGBW02009848
OWR49749.1
KQ460970
KPJ10151.1
+ More
KQ459597 KPI95207.1 KZ149956 PZC76465.1 KK852465 KDR23303.1 APGK01047104 KB741077 KB632384 ENN74134.1 ERL94265.1 KQ971307 EFA11582.2 KQ976398 KYM92756.1 NEVH01021574 PNF19617.1 KQ977259 KYN04551.1 GBHO01016926 JAG26678.1 ADTU01001646 ADTU01001647 ADTU01001648 ACPB03006188 ACPB03006189 GECZ01007650 JAS62119.1 GL888911 EGI57398.1 KQ982648 KYQ53213.1 GBYB01015189 JAG84956.1 QOIP01000011 RLU16646.1 GL434739 EFN74624.1 KK107078 EZA60201.1 KQ978713 KYN29072.1 KQ981979 KYN31367.1 DS235793 EEB17141.1 JXUM01061486 KQ562152 KXJ76555.1 CH477680 EAT37313.1 AXCN02000483 GBBI01003131 JAC15581.1 KJ512108 AID60331.1 AJWK01003104 UFQS01000716 UFQT01000716 SSX06386.1 SSX26740.1 AAAB01008960 EAA11233.5 CVRI01000035 CRK92900.1 APCN01002316 AXCM01006125 CM002911 KMY92016.1 KMY92017.1 CH954177 EDV59667.2 AE013599 AAF59230.2 AAS64900.1 CM000157 EDW89203.2 CH480816 EDW46779.1 GAKP01019583 JAC39369.1 CH933808 EDW09159.2 CH964154 EDW79846.1 CM000071 EAL25972.3 KRT02712.1 KRT02711.1 CH479223 EDW35029.1 CP012524 ALC42037.1 CH940648 EDW61524.2 CH902619 EDV36895.2 KPU76491.1 KPU76492.1 CH916367 EDW02212.1 AJVK01022752 AJVK01022753 KQ434825 KZC07370.1 PYGN01000240 PSN50388.1 UFQS01000742 UFQT01000742 SSX06522.1 SSX26871.1 ABLF02022531 ABLF02022532 ABLF02022533 ABLF02022534 ABLF02022535 ABLF02022536 ATLV01024451 ATLV01024452 KE525352 KFB51412.1 CAEY01000040 JXLN01006271 KPM03817.1 MUJZ01007520 OTF82633.1 NEDP02002422 OWF50829.1
KQ459597 KPI95207.1 KZ149956 PZC76465.1 KK852465 KDR23303.1 APGK01047104 KB741077 KB632384 ENN74134.1 ERL94265.1 KQ971307 EFA11582.2 KQ976398 KYM92756.1 NEVH01021574 PNF19617.1 KQ977259 KYN04551.1 GBHO01016926 JAG26678.1 ADTU01001646 ADTU01001647 ADTU01001648 ACPB03006188 ACPB03006189 GECZ01007650 JAS62119.1 GL888911 EGI57398.1 KQ982648 KYQ53213.1 GBYB01015189 JAG84956.1 QOIP01000011 RLU16646.1 GL434739 EFN74624.1 KK107078 EZA60201.1 KQ978713 KYN29072.1 KQ981979 KYN31367.1 DS235793 EEB17141.1 JXUM01061486 KQ562152 KXJ76555.1 CH477680 EAT37313.1 AXCN02000483 GBBI01003131 JAC15581.1 KJ512108 AID60331.1 AJWK01003104 UFQS01000716 UFQT01000716 SSX06386.1 SSX26740.1 AAAB01008960 EAA11233.5 CVRI01000035 CRK92900.1 APCN01002316 AXCM01006125 CM002911 KMY92016.1 KMY92017.1 CH954177 EDV59667.2 AE013599 AAF59230.2 AAS64900.1 CM000157 EDW89203.2 CH480816 EDW46779.1 GAKP01019583 JAC39369.1 CH933808 EDW09159.2 CH964154 EDW79846.1 CM000071 EAL25972.3 KRT02712.1 KRT02711.1 CH479223 EDW35029.1 CP012524 ALC42037.1 CH940648 EDW61524.2 CH902619 EDV36895.2 KPU76491.1 KPU76492.1 CH916367 EDW02212.1 AJVK01022752 AJVK01022753 KQ434825 KZC07370.1 PYGN01000240 PSN50388.1 UFQS01000742 UFQT01000742 SSX06522.1 SSX26871.1 ABLF02022531 ABLF02022532 ABLF02022533 ABLF02022534 ABLF02022535 ABLF02022536 ATLV01024451 ATLV01024452 KE525352 KFB51412.1 CAEY01000040 JXLN01006271 KPM03817.1 MUJZ01007520 OTF82633.1 NEDP02002422 OWF50829.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000027135
UP000019118
UP000030742
+ More
UP000007266 UP000078540 UP000235965 UP000078542 UP000005205 UP000015103 UP000002358 UP000007755 UP000075809 UP000279307 UP000000311 UP000053097 UP000078492 UP000078541 UP000005203 UP000009046 UP000069940 UP000249989 UP000008820 UP000075901 UP000075886 UP000075900 UP000092461 UP000075902 UP000007062 UP000183832 UP000075840 UP000075882 UP000075885 UP000076407 UP000075903 UP000091820 UP000075883 UP000075884 UP000075920 UP000079169 UP000095300 UP000095301 UP000075880 UP000192221 UP000008711 UP000000803 UP000002282 UP000001292 UP000069272 UP000009192 UP000007798 UP000001819 UP000008744 UP000092553 UP000008792 UP000007801 UP000001070 UP000092462 UP000076502 UP000245037 UP000007819 UP000030765 UP000015104 UP000242188 UP000261420 UP000076420
UP000007266 UP000078540 UP000235965 UP000078542 UP000005205 UP000015103 UP000002358 UP000007755 UP000075809 UP000279307 UP000000311 UP000053097 UP000078492 UP000078541 UP000005203 UP000009046 UP000069940 UP000249989 UP000008820 UP000075901 UP000075886 UP000075900 UP000092461 UP000075902 UP000007062 UP000183832 UP000075840 UP000075882 UP000075885 UP000076407 UP000075903 UP000091820 UP000075883 UP000075884 UP000075920 UP000079169 UP000095300 UP000095301 UP000075880 UP000192221 UP000008711 UP000000803 UP000002282 UP000001292 UP000069272 UP000009192 UP000007798 UP000001819 UP000008744 UP000092553 UP000008792 UP000007801 UP000001070 UP000092462 UP000076502 UP000245037 UP000007819 UP000030765 UP000015104 UP000242188 UP000261420 UP000076420
PRIDE
Pfam
Interpro
IPR001254
Trypsin_dom
+ More
IPR018114 TRYPSIN_HIS
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR020067 Frizzled_dom
IPR001190 SRCR
IPR036790 Frizzled_dom_sf
IPR036772 SRCR-like_dom_sf
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR017448 SRCR-like_dom
IPR023415 LDLR_class-A_CS
IPR036322 WD40_repeat_dom_sf
IPR000082 SEA_dom
IPR036364 SEA_dom_sf
IPR018114 TRYPSIN_HIS
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR020067 Frizzled_dom
IPR001190 SRCR
IPR036790 Frizzled_dom_sf
IPR036772 SRCR-like_dom_sf
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR017448 SRCR-like_dom
IPR023415 LDLR_class-A_CS
IPR036322 WD40_repeat_dom_sf
IPR000082 SEA_dom
IPR036364 SEA_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1V872
A0A212F7R4
A0A0N1IE78
A0A194PPH0
A0A2W1BS65
A0A067RJW0
+ More
N6T216 D6W7Q1 A0A195BXT6 A0A2J7PTF4 A0A195CV41 A0A0A9Y6G6 A0A158NQ02 T1I1Y6 A0A1B6GI55 K7IQW1 F4X817 A0A151WZ95 A0A0C9RF23 A0A3L8D8M9 E1ZW18 A0A026WWV7 A0A151JQ49 A0A195ETY3 A0A088ARG2 E0VUT5 A0A182GGD0 Q16SA2 A0A182S7G8 A0A182Q4B6 A0A023F359 A0A068F4F8 A0A182R978 A0A1B0C9Z5 A0A336KNG9 A0A182TY97 Q7Q554 A0A1J1I006 A0A182I4L0 A0A182LKB7 A0A182PDL4 A0A182X4L3 A0A182VBY8 A0A1A9X3N6 A0A182LTZ4 A0A182NI76 A0A182WBN6 A0A1S4ESL9 A0A1I8PR60 A0A1I8PRA4 A0A1I8MUQ7 A0A182J3Y1 A0A1W4VDJ1 A0A0J9R7H2 B3N9X1 A1Z709 B4P1R7 B4HR36 A0A182FEA1 A0A034VA32 B4KPP9 B4N657 Q28YJ3 A0A0R3NNY5 B4H8J4 A0A0M4EVS7 B4LJ88 B3MHB8 B4J7Y2 A0A1B0D2D8 A0A154P8C6 A0A2P8Z1M6 A0A336MC12 J9KVA0 A0A084WMG8 T1KG36 A0A131ZYS7 A0A1Y3BS63 A0A210QQ31 A0A3B4T7E3 A0A3B4T6Y7 A0A3B4T6V1 A0A3B4T6M8 A0A3B4T6L2 A0A2C9KCV1
N6T216 D6W7Q1 A0A195BXT6 A0A2J7PTF4 A0A195CV41 A0A0A9Y6G6 A0A158NQ02 T1I1Y6 A0A1B6GI55 K7IQW1 F4X817 A0A151WZ95 A0A0C9RF23 A0A3L8D8M9 E1ZW18 A0A026WWV7 A0A151JQ49 A0A195ETY3 A0A088ARG2 E0VUT5 A0A182GGD0 Q16SA2 A0A182S7G8 A0A182Q4B6 A0A023F359 A0A068F4F8 A0A182R978 A0A1B0C9Z5 A0A336KNG9 A0A182TY97 Q7Q554 A0A1J1I006 A0A182I4L0 A0A182LKB7 A0A182PDL4 A0A182X4L3 A0A182VBY8 A0A1A9X3N6 A0A182LTZ4 A0A182NI76 A0A182WBN6 A0A1S4ESL9 A0A1I8PR60 A0A1I8PRA4 A0A1I8MUQ7 A0A182J3Y1 A0A1W4VDJ1 A0A0J9R7H2 B3N9X1 A1Z709 B4P1R7 B4HR36 A0A182FEA1 A0A034VA32 B4KPP9 B4N657 Q28YJ3 A0A0R3NNY5 B4H8J4 A0A0M4EVS7 B4LJ88 B3MHB8 B4J7Y2 A0A1B0D2D8 A0A154P8C6 A0A2P8Z1M6 A0A336MC12 J9KVA0 A0A084WMG8 T1KG36 A0A131ZYS7 A0A1Y3BS63 A0A210QQ31 A0A3B4T7E3 A0A3B4T6Y7 A0A3B4T6V1 A0A3B4T6M8 A0A3B4T6L2 A0A2C9KCV1
PDB
4DGJ
E-value=1.91652e-39,
Score=406
Ontologies
GO
Topology
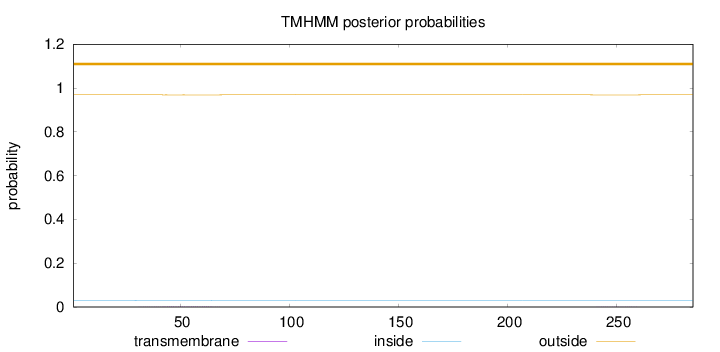
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0468
Exp number, first 60 AAs:
0.03551
Total prob of N-in:
0.03008
outside
1 - 285
Population Genetic Test Statistics
Pi
190.34667
Theta
16.956839
Tajima's D
0.755886
CLR
1.086007
CSRT
0.60301984900755
Interpretation
Uncertain
