Pre Gene Modal
BGIBMGA005748
Annotation
PREDICTED:_gamma-1-syntrophin_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.065
Sequence
CDS
ATGCCATTAAAGTTGGCTTTGATATCATACACCAGAGTGATCGAATTATTTTTTATTTCAGTTAATGGTGAATACATAACAGCCTGCAAACACGACGATGCTGTGAACATACTGAGGAACGCCGGTGATATCGTTGTACTAACTGTGAAGCATTATAGGGCTGCCACACCTTTCTTACAGAAGCAAGCTGACGGAGCATCAGATACAGACAGCAGCACCGGCGACGATCACCCATCGCTTCACAACAAGGTCGATGCGAATTGCAACTCGGAGAAGAGTTCAGAACACACCAATGGTGGTTGGCAGAGCGCTTGGCCTGATGAAGAATCGAGTAGACCGGGGAGTGCTACAGGGTCGGTGAGAGGCTCCCGGCCTCCCTCGGTGGCTTCGGATAGACACGATGTCACCATTAAGGAATGGGTCGACGTCGTATCAGTGCCATTAATGATGGGATACGTAACGCGATACGTCCTTGGAACGGATAAATTGAGACCTGGAGCTTTTGAGGTCCGAGGAGCTCGTCCGCATTTAACCACCGGAGTGATACACGTGGACGATCCCGTGGCGCTTTCTCACTGGCTGAAGAGCATTTCCGATAATATCGTGGGACTCACGAACTTACAGATGAAACTTTTTAACCGCCACCTGGCGGCTGGAGAGCGGATCGAGTACATGGGTTGGGTGCACGAGGGCGTGGTCACGGCCCCGCAGCCCTGGCAGTCCTATAGACCGAAGTTCTTGGTACTGAAAGGAACTGACGTCATGCTTTTTGACGTACCACCGATGAACATGACCGAGCTAGCGAAATGCCCAGAAAGCCTGAAAGTTTACCAGACCATGTTCAGGACTGTAAAAGAGAGCGAGACGGTGGACTGGAGGCAGCACTGCTTCTTGGTGCAGAGCAGCAGCGCCGGGGTCGGGACCCCAGGGCCGAGGTACTTCAGCGCGGAGACCAGACAGGAACTGCTGAGGGTCGAGGCCGCCTGGACAGCTAACATCGTCAACTCTGTTATAAGACTCGGGAAAAAGACATTCACGGTTGTGCATTCAGGACGTGCGGCCGGGCTTACGTTGGATTGGTCGGCGGGGTTCTCGCTCACCGAGGGCACACCCGGAGCGTCTCCCGTCTGGTCGTATAGGTTTTCGCAGTTAAGAGGTTCGAGTGACGATGGAAAATCGAAATTGAAACTCCACTTCCAAGACGCCGAAACGAAGGTCATTGAAACAAAAGAATTAGAATGTCAAATACTGCAGAGTCTATTGTTCTGCATGCACGCCTTCCTGACCGCAAAGGTGGCCTCCGTAGACCCGGCCTTCCTCGCGTCCATACAGCAGTCTAATTAA
Protein
MPLKLALISYTRVIELFFISVNGEYITACKHDDAVNILRNAGDIVVLTVKHYRAATPFLQKQADGASDTDSSTGDDHPSLHNKVDANCNSEKSSEHTNGGWQSAWPDEESSRPGSATGSVRGSRPPSVASDRHDVTIKEWVDVVSVPLMMGYVTRYVLGTDKLRPGAFEVRGARPHLTTGVIHVDDPVALSHWLKSISDNIVGLTNLQMKLFNRHLAAGERIEYMGWVHEGVVTAPQPWQSYRPKFLVLKGTDVMLFDVPPMNMTELAKCPESLKVYQTMFRTVKESETVDWRQHCFLVQSSSAGVGTPGPRYFSAETRQELLRVEAAWTANIVNSVIRLGKKTFTVVHSGRAAGLTLDWSAGFSLTEGTPGASPVWSYRFSQLRGSSDDGKSKLKLHFQDAETKVIETKELECQILQSLLFCMHAFLTAKVASVDPAFLASIQQSN
Summary
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
A0A2H1W791
A0A2W1BUY9
H9J8A6
A0A2H1WV25
A0A182FE98
A0A2M4BK76
+ More
B4J7G9 W8BHR8 A0A182QFJ9 A0A0A1X8M1 A0A182I4L2 A0A182X4L5 A0A0K8VH69 A0A1L8E3F6 A0A182R980 A0A182NI74 A0A182UYP0 A0A1Q3FVV2 A0A182Y2H4 A0A084WMG4 A0A023EUF5 B4KSI4 A0A0K8W9G5 B4LMF1 A0A1L8E3R3 T1E1X1 A0A182J981 A0A182PDL2 Q16GC9 Q7PPA0 B0X6I9 A0A3B0JQN0 B4GA94 Q291U2 A0A0L0BNX1 A0A182WBN4 A0A1W4UYC3 A0A1W4UYP1 B3MFQ4 A0A1I8PIY3 A0A0J9RFD3 E3CTN9 A0A0R1DY73 B4P657 A0A2M4CPD3 A0A0J9RED0 W5JMY3 Q8SX22 A1ZAH3 A0A0Q5VZR4 B3NPH5 B4MPH6 A0A1B0FKQ0 D3PFI3 D6W7Q2 A0A0T6AYZ3 A0A1J1HZD9 B4HSZ3 A0A1Y1LY32 N6TT16 A0A1W4X3S4 A0A0N0PBB7 B4QI03 A0A2A3E8X9 A0A087ZUY4 A0A1B6HUC8 T1ID62 A0A067RRM7 A0A023F148 A0A0L7QS01 A0A154PR79 A0A0C9RNC1 A0A0M8ZU42 A0A1B6D4W2 A0A336MS73 E2BYV7 A0A158NNW7 A0A182TGX4 A0A151I949 A0A195EX22 A0A026WQP4 A0A3L8DMP0 A0A336MQX6 A0A182LKB4 A7UUG4 A0A182MQS5 A0A182SQP7 A0A232F6G4 A0A195BRW9 E2AEK9 A0A1I8PJ89 A0A336N037 A0A0M4EV41 F4WV84 A0A2J7PTG2 A0A0A9YJY5 E0VUS8
B4J7G9 W8BHR8 A0A182QFJ9 A0A0A1X8M1 A0A182I4L2 A0A182X4L5 A0A0K8VH69 A0A1L8E3F6 A0A182R980 A0A182NI74 A0A182UYP0 A0A1Q3FVV2 A0A182Y2H4 A0A084WMG4 A0A023EUF5 B4KSI4 A0A0K8W9G5 B4LMF1 A0A1L8E3R3 T1E1X1 A0A182J981 A0A182PDL2 Q16GC9 Q7PPA0 B0X6I9 A0A3B0JQN0 B4GA94 Q291U2 A0A0L0BNX1 A0A182WBN4 A0A1W4UYC3 A0A1W4UYP1 B3MFQ4 A0A1I8PIY3 A0A0J9RFD3 E3CTN9 A0A0R1DY73 B4P657 A0A2M4CPD3 A0A0J9RED0 W5JMY3 Q8SX22 A1ZAH3 A0A0Q5VZR4 B3NPH5 B4MPH6 A0A1B0FKQ0 D3PFI3 D6W7Q2 A0A0T6AYZ3 A0A1J1HZD9 B4HSZ3 A0A1Y1LY32 N6TT16 A0A1W4X3S4 A0A0N0PBB7 B4QI03 A0A2A3E8X9 A0A087ZUY4 A0A1B6HUC8 T1ID62 A0A067RRM7 A0A023F148 A0A0L7QS01 A0A154PR79 A0A0C9RNC1 A0A0M8ZU42 A0A1B6D4W2 A0A336MS73 E2BYV7 A0A158NNW7 A0A182TGX4 A0A151I949 A0A195EX22 A0A026WQP4 A0A3L8DMP0 A0A336MQX6 A0A182LKB4 A7UUG4 A0A182MQS5 A0A182SQP7 A0A232F6G4 A0A195BRW9 E2AEK9 A0A1I8PJ89 A0A336N037 A0A0M4EV41 F4WV84 A0A2J7PTG2 A0A0A9YJY5 E0VUS8
Pubmed
28756777
19121390
17994087
24495485
25830018
25244985
+ More
24438588 24945155 18057021 24330624 17510324 12364791 14747013 17210077 15632085 23185243 26108605 22936249 17550304 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 28004739 23537049 26354079 24845553 25474469 20798317 21347285 24508170 30249741 20966253 28648823 21719571 25401762 20566863
24438588 24945155 18057021 24330624 17510324 12364791 14747013 17210077 15632085 23185243 26108605 22936249 17550304 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 28004739 23537049 26354079 24845553 25474469 20798317 21347285 24508170 30249741 20966253 28648823 21719571 25401762 20566863
EMBL
ODYU01006475
SOQ48344.1
KZ149956
PZC76466.1
BABH01027508
BABH01027509
+ More
BABH01027510 ODYU01011274 SOQ56910.1 GGFJ01004335 MBW53476.1 CH916367 EDW01093.1 GAMC01010073 GAMC01010072 JAB96483.1 AXCN02000484 GBXI01006830 JAD07462.1 APCN01002317 GDHF01014409 JAI37905.1 GFDF01000811 JAV13273.1 GFDL01003304 JAV31741.1 ATLV01024447 ATLV01024448 KE525352 KFB51408.1 GAPW01001021 JAC12577.1 CH933808 EDW09489.1 GDHF01004481 JAI47833.1 CH940648 EDW62046.2 KRF80401.1 KRF80402.1 KRF80403.1 GFDF01000810 JAV13274.1 GALA01001695 JAA93157.1 CH478293 EAT33296.1 AAAB01008960 EAA10804.4 DS232415 EDS41472.1 OUUW01000001 SPP73458.1 CH479181 EDW31846.1 CM000071 EAL25020.1 KRT01799.1 JRES01001695 KNC20914.1 CH902619 EDV37744.1 CM002911 KMY94284.1 BT125705 ADO33196.1 CM000158 KRK00202.1 EDW91907.2 GGFL01002951 MBW67129.1 KMY94285.1 KMY94286.1 ADMH02000584 ETN65737.1 AE013599 AY094895 AAF57987.2 AAM11248.1 AAM68501.1 BT132953 AAO41368.1 AEW43893.1 CH954179 KQS62449.1 KQS62450.1 EDV55742.2 CH963849 EDW74015.1 CCAG010001145 CCAG010001146 CCAG010001147 BT120389 ADD16469.1 KQ971307 EFA11248.1 LJIG01022578 KRT79831.1 CVRI01000035 CRK92898.1 CH480816 EDW48157.1 GEZM01046912 JAV76965.1 APGK01020125 KB740167 KB631669 ENN81198.1 ERL85165.1 KQ460970 KPJ10148.1 CM000362 EDX07374.1 KZ288345 PBC27649.1 GECU01029428 JAS78278.1 ACPB03022052 KK852465 KDR23305.1 GBBI01003470 JAC15242.1 KQ414766 KOC61402.1 KQ435007 KZC13620.1 GBYB01014992 JAG84759.1 KQ435845 KOX71185.1 GEDC01016567 GEDC01009604 JAS20731.1 JAS27694.1 UFQT01002209 SSX32930.1 GL451531 EFN79129.1 ADTU01021859 ADTU01021860 KQ978303 KYM95335.1 KQ981953 KYN32429.1 KK107128 EZA58350.1 QOIP01000006 RLU21690.1 UFQT01001726 SSX31539.1 EDO63942.1 AXCM01016171 NNAY01000802 OXU26434.1 KQ976417 KYM89760.1 GL438870 EFN68132.1 SSX31538.1 CP012524 ALC41675.1 GL888384 EGI61850.1 NEVH01021574 PNF19614.1 GBHO01010222 JAG33382.1 DS235793 EEB17134.1
BABH01027510 ODYU01011274 SOQ56910.1 GGFJ01004335 MBW53476.1 CH916367 EDW01093.1 GAMC01010073 GAMC01010072 JAB96483.1 AXCN02000484 GBXI01006830 JAD07462.1 APCN01002317 GDHF01014409 JAI37905.1 GFDF01000811 JAV13273.1 GFDL01003304 JAV31741.1 ATLV01024447 ATLV01024448 KE525352 KFB51408.1 GAPW01001021 JAC12577.1 CH933808 EDW09489.1 GDHF01004481 JAI47833.1 CH940648 EDW62046.2 KRF80401.1 KRF80402.1 KRF80403.1 GFDF01000810 JAV13274.1 GALA01001695 JAA93157.1 CH478293 EAT33296.1 AAAB01008960 EAA10804.4 DS232415 EDS41472.1 OUUW01000001 SPP73458.1 CH479181 EDW31846.1 CM000071 EAL25020.1 KRT01799.1 JRES01001695 KNC20914.1 CH902619 EDV37744.1 CM002911 KMY94284.1 BT125705 ADO33196.1 CM000158 KRK00202.1 EDW91907.2 GGFL01002951 MBW67129.1 KMY94285.1 KMY94286.1 ADMH02000584 ETN65737.1 AE013599 AY094895 AAF57987.2 AAM11248.1 AAM68501.1 BT132953 AAO41368.1 AEW43893.1 CH954179 KQS62449.1 KQS62450.1 EDV55742.2 CH963849 EDW74015.1 CCAG010001145 CCAG010001146 CCAG010001147 BT120389 ADD16469.1 KQ971307 EFA11248.1 LJIG01022578 KRT79831.1 CVRI01000035 CRK92898.1 CH480816 EDW48157.1 GEZM01046912 JAV76965.1 APGK01020125 KB740167 KB631669 ENN81198.1 ERL85165.1 KQ460970 KPJ10148.1 CM000362 EDX07374.1 KZ288345 PBC27649.1 GECU01029428 JAS78278.1 ACPB03022052 KK852465 KDR23305.1 GBBI01003470 JAC15242.1 KQ414766 KOC61402.1 KQ435007 KZC13620.1 GBYB01014992 JAG84759.1 KQ435845 KOX71185.1 GEDC01016567 GEDC01009604 JAS20731.1 JAS27694.1 UFQT01002209 SSX32930.1 GL451531 EFN79129.1 ADTU01021859 ADTU01021860 KQ978303 KYM95335.1 KQ981953 KYN32429.1 KK107128 EZA58350.1 QOIP01000006 RLU21690.1 UFQT01001726 SSX31539.1 EDO63942.1 AXCM01016171 NNAY01000802 OXU26434.1 KQ976417 KYM89760.1 GL438870 EFN68132.1 SSX31538.1 CP012524 ALC41675.1 GL888384 EGI61850.1 NEVH01021574 PNF19614.1 GBHO01010222 JAG33382.1 DS235793 EEB17134.1
Proteomes
UP000005204
UP000069272
UP000001070
UP000075886
UP000075840
UP000076407
+ More
UP000075900 UP000075884 UP000075903 UP000076408 UP000030765 UP000009192 UP000008792 UP000075880 UP000075885 UP000008820 UP000007062 UP000002320 UP000268350 UP000008744 UP000001819 UP000037069 UP000075920 UP000192221 UP000007801 UP000095300 UP000002282 UP000000673 UP000000803 UP000008711 UP000007798 UP000092444 UP000007266 UP000183832 UP000001292 UP000019118 UP000030742 UP000192223 UP000053240 UP000000304 UP000242457 UP000005203 UP000015103 UP000027135 UP000053825 UP000076502 UP000053105 UP000008237 UP000005205 UP000075902 UP000078542 UP000078541 UP000053097 UP000279307 UP000075882 UP000075883 UP000075901 UP000215335 UP000078540 UP000000311 UP000092553 UP000007755 UP000235965 UP000009046
UP000075900 UP000075884 UP000075903 UP000076408 UP000030765 UP000009192 UP000008792 UP000075880 UP000075885 UP000008820 UP000007062 UP000002320 UP000268350 UP000008744 UP000001819 UP000037069 UP000075920 UP000192221 UP000007801 UP000095300 UP000002282 UP000000673 UP000000803 UP000008711 UP000007798 UP000092444 UP000007266 UP000183832 UP000001292 UP000019118 UP000030742 UP000192223 UP000053240 UP000000304 UP000242457 UP000005203 UP000015103 UP000027135 UP000053825 UP000076502 UP000053105 UP000008237 UP000005205 UP000075902 UP000078542 UP000078541 UP000053097 UP000279307 UP000075882 UP000075883 UP000075901 UP000215335 UP000078540 UP000000311 UP000092553 UP000007755 UP000235965 UP000009046
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1W791
A0A2W1BUY9
H9J8A6
A0A2H1WV25
A0A182FE98
A0A2M4BK76
+ More
B4J7G9 W8BHR8 A0A182QFJ9 A0A0A1X8M1 A0A182I4L2 A0A182X4L5 A0A0K8VH69 A0A1L8E3F6 A0A182R980 A0A182NI74 A0A182UYP0 A0A1Q3FVV2 A0A182Y2H4 A0A084WMG4 A0A023EUF5 B4KSI4 A0A0K8W9G5 B4LMF1 A0A1L8E3R3 T1E1X1 A0A182J981 A0A182PDL2 Q16GC9 Q7PPA0 B0X6I9 A0A3B0JQN0 B4GA94 Q291U2 A0A0L0BNX1 A0A182WBN4 A0A1W4UYC3 A0A1W4UYP1 B3MFQ4 A0A1I8PIY3 A0A0J9RFD3 E3CTN9 A0A0R1DY73 B4P657 A0A2M4CPD3 A0A0J9RED0 W5JMY3 Q8SX22 A1ZAH3 A0A0Q5VZR4 B3NPH5 B4MPH6 A0A1B0FKQ0 D3PFI3 D6W7Q2 A0A0T6AYZ3 A0A1J1HZD9 B4HSZ3 A0A1Y1LY32 N6TT16 A0A1W4X3S4 A0A0N0PBB7 B4QI03 A0A2A3E8X9 A0A087ZUY4 A0A1B6HUC8 T1ID62 A0A067RRM7 A0A023F148 A0A0L7QS01 A0A154PR79 A0A0C9RNC1 A0A0M8ZU42 A0A1B6D4W2 A0A336MS73 E2BYV7 A0A158NNW7 A0A182TGX4 A0A151I949 A0A195EX22 A0A026WQP4 A0A3L8DMP0 A0A336MQX6 A0A182LKB4 A7UUG4 A0A182MQS5 A0A182SQP7 A0A232F6G4 A0A195BRW9 E2AEK9 A0A1I8PJ89 A0A336N037 A0A0M4EV41 F4WV84 A0A2J7PTG2 A0A0A9YJY5 E0VUS8
B4J7G9 W8BHR8 A0A182QFJ9 A0A0A1X8M1 A0A182I4L2 A0A182X4L5 A0A0K8VH69 A0A1L8E3F6 A0A182R980 A0A182NI74 A0A182UYP0 A0A1Q3FVV2 A0A182Y2H4 A0A084WMG4 A0A023EUF5 B4KSI4 A0A0K8W9G5 B4LMF1 A0A1L8E3R3 T1E1X1 A0A182J981 A0A182PDL2 Q16GC9 Q7PPA0 B0X6I9 A0A3B0JQN0 B4GA94 Q291U2 A0A0L0BNX1 A0A182WBN4 A0A1W4UYC3 A0A1W4UYP1 B3MFQ4 A0A1I8PIY3 A0A0J9RFD3 E3CTN9 A0A0R1DY73 B4P657 A0A2M4CPD3 A0A0J9RED0 W5JMY3 Q8SX22 A1ZAH3 A0A0Q5VZR4 B3NPH5 B4MPH6 A0A1B0FKQ0 D3PFI3 D6W7Q2 A0A0T6AYZ3 A0A1J1HZD9 B4HSZ3 A0A1Y1LY32 N6TT16 A0A1W4X3S4 A0A0N0PBB7 B4QI03 A0A2A3E8X9 A0A087ZUY4 A0A1B6HUC8 T1ID62 A0A067RRM7 A0A023F148 A0A0L7QS01 A0A154PR79 A0A0C9RNC1 A0A0M8ZU42 A0A1B6D4W2 A0A336MS73 E2BYV7 A0A158NNW7 A0A182TGX4 A0A151I949 A0A195EX22 A0A026WQP4 A0A3L8DMP0 A0A336MQX6 A0A182LKB4 A7UUG4 A0A182MQS5 A0A182SQP7 A0A232F6G4 A0A195BRW9 E2AEK9 A0A1I8PJ89 A0A336N037 A0A0M4EV41 F4WV84 A0A2J7PTG2 A0A0A9YJY5 E0VUS8
PDB
1Z87
E-value=0.0107819,
Score=92
Ontologies
GO
PANTHER
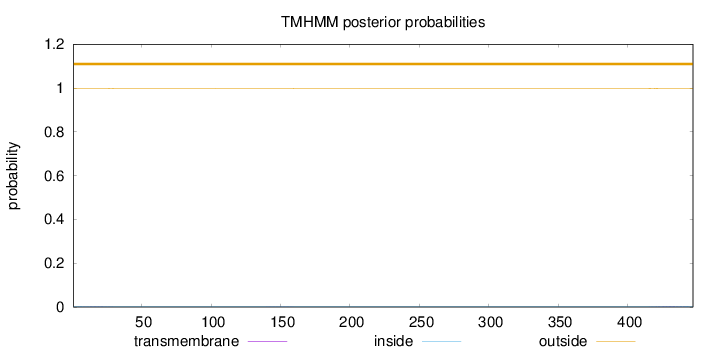
Topology
Length:
447
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0901
Exp number, first 60 AAs:
0.02872
Total prob of N-in:
0.00250
outside
1 - 447
Population Genetic Test Statistics
Pi
237.294526
Theta
186.046883
Tajima's D
0.341249
CLR
0.608193
CSRT
0.471376431178441
Interpretation
Uncertain
