Gene
KWMTBOMO03055
Pre Gene Modal
BGIBMGA005747
Annotation
PREDICTED:_Down_syndrome_cell_adhesion_molecule-like_protein_Dscam2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.792 PlasmaMembrane Reliability : 1.095
Sequence
CDS
ATGTCTGAAGTATCGCCAAAGTGGGTATTGGAGCCCAAGGATGAAGCTGTTTTGGCTGGCGAGTCTCTTTCTATTCACTGTCAAACGACCGGATCTCCAACGCCACAGGTCACGTGGTTAAAGCAGAGGGGTGGGACTTCGTCAGCTGATTACGCACCTCTGGTAAACCTCGGTGGCAGGTTTCAATTCTATCTGAACGGCACGTTGTGGATCGAGTCAGCCCTGGCCTACGACGAGGGAAATTATATGTGCAAATCGGAAAATGGTGTGGGCACCGCACTCACTAAAATCATTTTCGTTGCTGTCAACGAGCCAGCCAGGTTTGAAGTGACGTCACACAATATGACGGCGCGTCGTGACGCACCCGTGACGCTGTCTTGTGACGTCCGCGGTGACAGTCCTATACGCGTCACGTGGGCTCACAACACGAATCACTTAGATCTTAACACTTATAGGCTAAGTATCTCCGAAGCAAAGACTGATAGTGGTTTAAGGTCGCAACTGTACATCAGCAGAACAGACAGACAGGACTCCGGAATCTACAAGTGCCAGGCCGTCAACGCGTATGGTCATAGTGACCATTACATTTATTTATCCGTCCAAGAAAAACCTGACACGCCCCATTCACTGTCCGTGATCGAGATCCTGAGTCGGGCCGTTCGTTTGTCCTGGAGTCAGGGCTTCGATGGCAACAGCCCGCTCATGGGATACACGCTGCAGTACGCCGTGCTATCCACACAAGGCGCTATACGGGTCAACCAATGGGATTCTGCGCATAGCGTTAACGTTTCCGTCCATGGAGCTTCCCAGTCTCATGTTACATTGACGAAGAAAGGTGACATTCACTACGAAGCTCTACTCAGCAGCTTGAAGCCTCACACGGCGTACATGATTCGGATAGCAGCCATCAACCAAATAGACAGAAGTACCTTCACTGAACCAGTTGTCGTGAAAACACAAGAAGAAGCACCATCAGAGCCGCCTACGAATGTCCACGTGACTCCTGGAGGTCCTGGCGAGCTGCACGTGTCCTGGCGGCCCCCTACCCGTGAGGCGTGGCATGGAGAACTTCTCGGATACTCGGTAACATGCACCGAAGTGAACCCGGCTGGTCCATCAATGAGGAATGCTTCAAGAACCTTAACAGTTAATGGTTGGTCGGCCACTGAACTGAGTTTGTCTGCACTGCGAAAGTTCACGCGGTACGAAGTCCGGGTACGAGCTTTCAATGGAGTGGCAGCAGGACCGGCTTCTGCTCCTGTATCAGCTACTACTCTTGAGGGGGTTCCAGAATCCCATCCCACAAGAGTTTCCTGTGCGGCTCTATCGTCATCCAGCATGAAGGTGTCTTGGAACCCTCCTCCGATCGGACAACATGGAGGTCTCATTCAGGGCTATAAGGTTATCTATGCACCGCTTTCAACGGACCATGAAGATGTCGGTGAAATTAAGCGTGTGACCACAACTGACACCTATCTGCACACTCTAAGGAAATACACTAACTACTCCGTACAAGTTTTGGCTTTCACGAATGCCGGTGATGGAAAACGGAGTCCTCCTATCCACTGTATGACTGAGGAAGATGTACCATCAGCTCCTGAAAACATCAAAGCTCTCGCGTATTCCATTGACTCGGTTCTGGTGAGCTGGCTACCCCCGGTTCATCCAAACGGTGTTATTTCACATTACACAGTTTACTACCGAGAAGCCGGACGATTAGGCAAGCACTCCAGCTTCACTGTGTCAGCTGATAAGCAAACGGATGCGGAACTAATGTTCCAAGTGCGAAATTTAGCTGAAAATCAGCTTTACGAATTCTGGGTGTCCGCCACCACTGGTTCCGGAGAAGGGGAGTCCACGCTTGTAGTCGGCCAGGGACCTAGCTCTAGAATTCCAGCGCGCATCGCTTCGTTCGGGCGTTGGGTATCTGTGGGAGTTGGTCAGTCGCTGCTGCTTTTGTGCACGTGCGTGGGCTCGCCGACGCCGCGCTCGCGGTGGACACATGCGCGGTCGCCCGTCACGCACCACGCCTTCTACCAGGTCACGCACCGAGGACATCTGCATATACGAGAAGTAAACGAGCAGTCCTCAGGAAACTTCACATGTACAGCGAGCAACACGATAGGAGAGGACTCCATAGTGTACGTAGTGGCAGCCATCCTTCCTCCAGCCGCGCCATCCCTAAAGCTCCAGTACACCACCACGTCTAGTATAAAGCTGCACTGGCAACTACCGAGCGAAAATACCGAACCGATATTGGGGTATCTATTACACTACAAACAAGCATCTGAAGCCGAATGGCATACTGTGGAACTATCTCCTGAAGTTACATCGTACACTTTGGACATGCTGAAATGTGGCACGACGTACAACGCGAAGATACAAGCCAGAAATAAGATATCTATAGGACCACCCAGTGAGATCCTCACGGCCACTACCAGAGGAGGACGTCCAAAGCCTCCAAAGCCTGAAGACCACGTGCATATGAATTCCACAGCGATCAAAATAAACTTCTACGCTTGGCGTGACGGCGGCTGCCCCATTCTCGGCTTCAGAGTCTCTTACCGGAAAGCTGGAACAGAACATTGGATAAAGGTCGGGGATGGACTGAGTTCAGCCGGCCACGTGATCGGCTCTCTGACACCCGGAACATGGTATGAGGTTTGCGTGGAAGCCTACAGCGACGCTGGACCCGAACGTGTGACCCTACACGTCGATACACACACCTTGGCCGGAGGTCGCATTCCACCACTTAAGCCTTCATCACCGCTGTCGGGTAGCTCGCGGCAAAGCTCGCTACGAACCGTGCTGGTGTCTTGTGTGGCCAGCGGAGTCGTCATTGTCGTGACGTTGGCAGCGATCATCTGTCTAATACATGGAAAAAAGAAATTCTTCTGCCTTTCAAGTGACCACTACATGAGAGATGATAGAAAACTGAGCGAGAGTAACGAAGCGGAAAGAGAGAAGCTAAGAGAAGGACAGAAACTATACTCGTCGTCTTCTATCAATGGAAACGAAAAGTCAAACGATGACTCTTCAGCCGAACTGTACGAGATAAGTCCGTACGCGACTTTCGGCGTGTCGGCGCACTCGCTCCAGTTCCGCACGCTGGCGCGCCGAGAGGACGACGCGGCGCCCCCGCACCGCCGCCGCCACCGGGCCTGCGACCACTATCGATACGATGAATCGAGCCTCTCTAAATGCTCGACAGTGGAGGCGCGGCACCGCATGCGCGCGGCGCCCGCCGCCCCCTGGAGGGACGCGCACACCGACTCGGACGACAACAGCGATAGTGCCGCCAACACCACTACTAAGGGGTCGGGCACATCGCCGTGTGGCAGCGCTTACGGCAGCGGCAGACGGAGCGCCCGCACCACCAGCAGCGGAGGTCCAAATGGGAGCGACGGTCATTCAGACAGTTTCGCCCCCGTCCCCGCTGATATATCTTCTTTAATCGACAAATACCAACAGAGAAAAGAACAAGAACGTCGAGAATGTACAATACACGTGTAG
Protein
MSEVSPKWVLEPKDEAVLAGESLSIHCQTTGSPTPQVTWLKQRGGTSSADYAPLVNLGGRFQFYLNGTLWIESALAYDEGNYMCKSENGVGTALTKIIFVAVNEPARFEVTSHNMTARRDAPVTLSCDVRGDSPIRVTWAHNTNHLDLNTYRLSISEAKTDSGLRSQLYISRTDRQDSGIYKCQAVNAYGHSDHYIYLSVQEKPDTPHSLSVIEILSRAVRLSWSQGFDGNSPLMGYTLQYAVLSTQGAIRVNQWDSAHSVNVSVHGASQSHVTLTKKGDIHYEALLSSLKPHTAYMIRIAAINQIDRSTFTEPVVVKTQEEAPSEPPTNVHVTPGGPGELHVSWRPPTREAWHGELLGYSVTCTEVNPAGPSMRNASRTLTVNGWSATELSLSALRKFTRYEVRVRAFNGVAAGPASAPVSATTLEGVPESHPTRVSCAALSSSSMKVSWNPPPIGQHGGLIQGYKVIYAPLSTDHEDVGEIKRVTTTDTYLHTLRKYTNYSVQVLAFTNAGDGKRSPPIHCMTEEDVPSAPENIKALAYSIDSVLVSWLPPVHPNGVISHYTVYYREAGRLGKHSSFTVSADKQTDAELMFQVRNLAENQLYEFWVSATTGSGEGESTLVVGQGPSSRIPARIASFGRWVSVGVGQSLLLLCTCVGSPTPRSRWTHARSPVTHHAFYQVTHRGHLHIREVNEQSSGNFTCTASNTIGEDSIVYVVAAILPPAAPSLKLQYTTTSSIKLHWQLPSENTEPILGYLLHYKQASEAEWHTVELSPEVTSYTLDMLKCGTTYNAKIQARNKISIGPPSEILTATTRGGRPKPPKPEDHVHMNSTAIKINFYAWRDGGCPILGFRVSYRKAGTEHWIKVGDGLSSAGHVIGSLTPGTWYEVCVEAYSDAGPERVTLHVDTHTLAGGRIPPLKPSSPLSGSSRQSSLRTVLVSCVASGVVIVVTLAAIICLIHGKKKFFCLSSDHYMRDDRKLSESNEAEREKLREGQKLYSSSSINGNEKSNDDSSAELYEISPYATFGVSAHSLQFRTLARREDDAAPPHRRRHRACDHYRYDESSLSKCSTVEARHRMRAAPAAPWRDAHTDSDDNSDSAANTTTKGSGTSPCGSAYGSGRRSARTTSSGGPNGSDGHSDSFAPVPADISSLIDKYQQRKEQERRECTIHV
Summary
Uniprot
H9J8A5
A0A2H1VAH3
A0A2W1BND6
A0A2A4J5Z6
A0A212FEX3
A0A0N1PFK5
+ More
A0A194PP52 A0A0L7LB63 A0A139WPD8 A0A139WPP7 A0A139WPF3 A0A195BJQ9 F4W742 A0A151IA46 A0A151JRW6 A0A158NJC3 A0A195EYM2 A0A088APY9 A0A0L7R8H0 A0A0N0BC89 A0A3L8DRQ5 E2A3Y9 E9J1T2 K7J3G0 A0A232F8U8 A0A1J1HP71 A0A0T6AZU3 A0A0L0C0J5 B4N9U7 A0A026VYJ5 A0A1B0FNH0 A0A1I8Q177 A0A1I8Q183 A0A1I8Q151 A0A1B0BV85 A0A0P8XZR3 A0A1W4UT57 A0A1W4V5H6 B3LVQ3 A0A1W4USR1 A0A0Q9X715 W8BHF6 B4K7M7 A0A0Q9X218 A0A1B0CHP3 A0A336KKP0 A0A3B0KAQ1 A0A336M8I3 A0A0Q9WGA5 A0A0R3NK49 Q29A88 A0A0R3NGP1 A0A0R3NGB7 B4LY43 B4G2L9 A0A0R3NG76 A0A0Q9WFX9 A0A0M4ETQ6 A0A2J7PQK5 A0A2J7PQK1 A0A2J7PQP1 A0A0R1E4W5 A8JR35 A0A0R1E506 A0A0R1E3C0 Q7KSE9 Q9VEJ5 B4PVB9 B4IBG9 A0A2J7PQK3 A0A182FCX3 A0A1W7R6Y5 A0A1A9ZZV3 A0A182QYC3 A0A084WCP8 A0A182VI12 A0A182YC33 A0A154NY75 W5JR02 A0A182J4M0 N6TSR9 U4U3K5 B4JTV0 A0A182RKW3 A0A182LBP1 B3P3D4 Q16QI5 A0A1S4FT36 Q7Q9I6 A0A2H1VFK8 A0A182U083 A0A2W1BN39 A0A182P8T2 A0A2A4JM43 A0A182WK19 A0A2J7PZP0 A0A0L7LAQ9 A0A212F7U9 W8AYU6
A0A194PP52 A0A0L7LB63 A0A139WPD8 A0A139WPP7 A0A139WPF3 A0A195BJQ9 F4W742 A0A151IA46 A0A151JRW6 A0A158NJC3 A0A195EYM2 A0A088APY9 A0A0L7R8H0 A0A0N0BC89 A0A3L8DRQ5 E2A3Y9 E9J1T2 K7J3G0 A0A232F8U8 A0A1J1HP71 A0A0T6AZU3 A0A0L0C0J5 B4N9U7 A0A026VYJ5 A0A1B0FNH0 A0A1I8Q177 A0A1I8Q183 A0A1I8Q151 A0A1B0BV85 A0A0P8XZR3 A0A1W4UT57 A0A1W4V5H6 B3LVQ3 A0A1W4USR1 A0A0Q9X715 W8BHF6 B4K7M7 A0A0Q9X218 A0A1B0CHP3 A0A336KKP0 A0A3B0KAQ1 A0A336M8I3 A0A0Q9WGA5 A0A0R3NK49 Q29A88 A0A0R3NGP1 A0A0R3NGB7 B4LY43 B4G2L9 A0A0R3NG76 A0A0Q9WFX9 A0A0M4ETQ6 A0A2J7PQK5 A0A2J7PQK1 A0A2J7PQP1 A0A0R1E4W5 A8JR35 A0A0R1E506 A0A0R1E3C0 Q7KSE9 Q9VEJ5 B4PVB9 B4IBG9 A0A2J7PQK3 A0A182FCX3 A0A1W7R6Y5 A0A1A9ZZV3 A0A182QYC3 A0A084WCP8 A0A182VI12 A0A182YC33 A0A154NY75 W5JR02 A0A182J4M0 N6TSR9 U4U3K5 B4JTV0 A0A182RKW3 A0A182LBP1 B3P3D4 Q16QI5 A0A1S4FT36 Q7Q9I6 A0A2H1VFK8 A0A182U083 A0A2W1BN39 A0A182P8T2 A0A2A4JM43 A0A182WK19 A0A2J7PZP0 A0A0L7LAQ9 A0A212F7U9 W8AYU6
Pubmed
19121390
28756777
22118469
26354079
26227816
18362917
+ More
19820115 21719571 21347285 30249741 20798317 21282665 20075255 28648823 26108605 17994087 24508170 24495485 15632085 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24438588 25244985 20920257 23761445 23537049 20966253 17510324 12364791
19820115 21719571 21347285 30249741 20798317 21282665 20075255 28648823 26108605 17994087 24508170 24495485 15632085 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24438588 25244985 20920257 23761445 23537049 20966253 17510324 12364791
EMBL
BABH01027510
BABH01027511
BABH01027512
BABH01027513
ODYU01001341
SOQ37392.1
+ More
KZ149956 PZC76468.1 NWSH01002784 PCG67537.1 AGBW02008880 OWR52305.1 KQ460970 KPJ10147.1 KQ459597 KPI95211.1 JTDY01001854 KOB72718.1 KQ971307 KYB29804.1 KYB29801.1 KYB29803.1 KQ976453 KYM85408.1 GL887813 EGI69926.1 KQ978255 KYM95887.1 KQ978579 KYN30034.1 ADTU01017524 ADTU01017525 ADTU01017526 ADTU01017527 ADTU01017528 ADTU01017529 ADTU01017530 ADTU01017531 ADTU01017532 ADTU01017533 KQ981920 KYN32984.1 KQ414632 KOC67128.1 KQ435922 KOX68634.1 QOIP01000005 RLU23107.1 GL436519 EFN71841.1 GL767674 EFZ13261.1 NNAY01000640 OXU27246.1 CVRI01000006 CRK88017.1 LJIG01022460 KRT80459.1 JRES01001161 KNC24949.1 CH964232 EDW80662.2 KK107570 EZA48842.1 CCAG010010693 CCAG010010694 CCAG010010695 CCAG010010696 CCAG010010697 JXJN01021159 JXJN01021160 CH902617 KPU80416.1 EDV43677.1 CH933806 KRG01530.1 GAMC01005845 JAC00711.1 EDW15371.2 KRG01529.1 AJWK01012559 UFQS01000550 UFQT01000550 SSX04801.1 SSX25164.1 OUUW01000005 SPP80648.1 SSX04800.1 SSX25163.1 CH940650 KRF83565.1 CM000070 KRT00047.1 EAL27462.3 KRT00044.1 KRT00043.1 EDW67931.1 CH479179 EDW24064.1 KRT00045.1 KRT00046.1 KRT00048.1 KRT00049.1 KRF83564.1 CP012526 ALC46292.1 NEVH01022635 PNF18619.1 PNF18614.1 PNF18616.1 CM000160 KRK02722.1 AE014297 ABW08696.1 KRK02724.1 KRK02723.1 AAS65164.2 AAF55426.3 EDW95788.2 KRK02725.1 CH480827 EDW44727.1 PNF18615.1 GEHC01000744 JAV46901.1 AXCN02001019 AXCN02001020 ATLV01022806 ATLV01022807 ATLV01022808 ATLV01022809 ATLV01022810 ATLV01022811 KE525337 KFB47992.1 KQ434782 KZC04573.1 ADMH02000362 ETN66827.1 APGK01020326 APGK01020327 APGK01020328 KB740193 ENN81073.1 KB631669 ERL85186.1 CH916374 EDV91529.1 CH954181 EDV48714.1 CH477747 EAT36655.1 AAAB01008900 EAA09501.4 ODYU01002306 SOQ39628.1 PZC76469.1 NWSH01001004 PCG73155.1 NEVH01020333 PNF21807.1 JTDY01001907 KOB72568.1 AGBW02009825 OWR49816.1 GAMC01020469 JAB86086.1
KZ149956 PZC76468.1 NWSH01002784 PCG67537.1 AGBW02008880 OWR52305.1 KQ460970 KPJ10147.1 KQ459597 KPI95211.1 JTDY01001854 KOB72718.1 KQ971307 KYB29804.1 KYB29801.1 KYB29803.1 KQ976453 KYM85408.1 GL887813 EGI69926.1 KQ978255 KYM95887.1 KQ978579 KYN30034.1 ADTU01017524 ADTU01017525 ADTU01017526 ADTU01017527 ADTU01017528 ADTU01017529 ADTU01017530 ADTU01017531 ADTU01017532 ADTU01017533 KQ981920 KYN32984.1 KQ414632 KOC67128.1 KQ435922 KOX68634.1 QOIP01000005 RLU23107.1 GL436519 EFN71841.1 GL767674 EFZ13261.1 NNAY01000640 OXU27246.1 CVRI01000006 CRK88017.1 LJIG01022460 KRT80459.1 JRES01001161 KNC24949.1 CH964232 EDW80662.2 KK107570 EZA48842.1 CCAG010010693 CCAG010010694 CCAG010010695 CCAG010010696 CCAG010010697 JXJN01021159 JXJN01021160 CH902617 KPU80416.1 EDV43677.1 CH933806 KRG01530.1 GAMC01005845 JAC00711.1 EDW15371.2 KRG01529.1 AJWK01012559 UFQS01000550 UFQT01000550 SSX04801.1 SSX25164.1 OUUW01000005 SPP80648.1 SSX04800.1 SSX25163.1 CH940650 KRF83565.1 CM000070 KRT00047.1 EAL27462.3 KRT00044.1 KRT00043.1 EDW67931.1 CH479179 EDW24064.1 KRT00045.1 KRT00046.1 KRT00048.1 KRT00049.1 KRF83564.1 CP012526 ALC46292.1 NEVH01022635 PNF18619.1 PNF18614.1 PNF18616.1 CM000160 KRK02722.1 AE014297 ABW08696.1 KRK02724.1 KRK02723.1 AAS65164.2 AAF55426.3 EDW95788.2 KRK02725.1 CH480827 EDW44727.1 PNF18615.1 GEHC01000744 JAV46901.1 AXCN02001019 AXCN02001020 ATLV01022806 ATLV01022807 ATLV01022808 ATLV01022809 ATLV01022810 ATLV01022811 KE525337 KFB47992.1 KQ434782 KZC04573.1 ADMH02000362 ETN66827.1 APGK01020326 APGK01020327 APGK01020328 KB740193 ENN81073.1 KB631669 ERL85186.1 CH916374 EDV91529.1 CH954181 EDV48714.1 CH477747 EAT36655.1 AAAB01008900 EAA09501.4 ODYU01002306 SOQ39628.1 PZC76469.1 NWSH01001004 PCG73155.1 NEVH01020333 PNF21807.1 JTDY01001907 KOB72568.1 AGBW02009825 OWR49816.1 GAMC01020469 JAB86086.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000007266 UP000078540 UP000007755 UP000078542 UP000078492 UP000005205 UP000078541 UP000005203 UP000053825 UP000053105 UP000279307 UP000000311 UP000002358 UP000215335 UP000183832 UP000037069 UP000007798 UP000053097 UP000092444 UP000095300 UP000092460 UP000007801 UP000192221 UP000009192 UP000092461 UP000268350 UP000008792 UP000001819 UP000008744 UP000092553 UP000235965 UP000002282 UP000000803 UP000001292 UP000069272 UP000092445 UP000075886 UP000030765 UP000075903 UP000076408 UP000076502 UP000000673 UP000075880 UP000019118 UP000030742 UP000001070 UP000075900 UP000075882 UP000008711 UP000008820 UP000007062 UP000075902 UP000075885 UP000075920
UP000007266 UP000078540 UP000007755 UP000078542 UP000078492 UP000005205 UP000078541 UP000005203 UP000053825 UP000053105 UP000279307 UP000000311 UP000002358 UP000215335 UP000183832 UP000037069 UP000007798 UP000053097 UP000092444 UP000095300 UP000092460 UP000007801 UP000192221 UP000009192 UP000092461 UP000268350 UP000008792 UP000001819 UP000008744 UP000092553 UP000235965 UP000002282 UP000000803 UP000001292 UP000069272 UP000092445 UP000075886 UP000030765 UP000075903 UP000076408 UP000076502 UP000000673 UP000075880 UP000019118 UP000030742 UP000001070 UP000075900 UP000075882 UP000008711 UP000008820 UP000007062 UP000075902 UP000075885 UP000075920
Pfam
Interpro
IPR013106
Ig_V-set
+ More
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR027413 GROEL-like_equatorial_sf
IPR013151 Immunoglobulin
IPR018379 BEN_domain
IPR013087 Znf_C2H2_type
IPR003890 MIF4G-like_typ-3
IPR016021 MIF4-like_sf
IPR016024 ARM-type_fold
IPR003006 Ig/MHC_CS
IPR021012 Dscam_C
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR027413 GROEL-like_equatorial_sf
IPR013151 Immunoglobulin
IPR018379 BEN_domain
IPR013087 Znf_C2H2_type
IPR003890 MIF4G-like_typ-3
IPR016021 MIF4-like_sf
IPR016024 ARM-type_fold
IPR003006 Ig/MHC_CS
IPR021012 Dscam_C
Gene 3D
CDD
ProteinModelPortal
H9J8A5
A0A2H1VAH3
A0A2W1BND6
A0A2A4J5Z6
A0A212FEX3
A0A0N1PFK5
+ More
A0A194PP52 A0A0L7LB63 A0A139WPD8 A0A139WPP7 A0A139WPF3 A0A195BJQ9 F4W742 A0A151IA46 A0A151JRW6 A0A158NJC3 A0A195EYM2 A0A088APY9 A0A0L7R8H0 A0A0N0BC89 A0A3L8DRQ5 E2A3Y9 E9J1T2 K7J3G0 A0A232F8U8 A0A1J1HP71 A0A0T6AZU3 A0A0L0C0J5 B4N9U7 A0A026VYJ5 A0A1B0FNH0 A0A1I8Q177 A0A1I8Q183 A0A1I8Q151 A0A1B0BV85 A0A0P8XZR3 A0A1W4UT57 A0A1W4V5H6 B3LVQ3 A0A1W4USR1 A0A0Q9X715 W8BHF6 B4K7M7 A0A0Q9X218 A0A1B0CHP3 A0A336KKP0 A0A3B0KAQ1 A0A336M8I3 A0A0Q9WGA5 A0A0R3NK49 Q29A88 A0A0R3NGP1 A0A0R3NGB7 B4LY43 B4G2L9 A0A0R3NG76 A0A0Q9WFX9 A0A0M4ETQ6 A0A2J7PQK5 A0A2J7PQK1 A0A2J7PQP1 A0A0R1E4W5 A8JR35 A0A0R1E506 A0A0R1E3C0 Q7KSE9 Q9VEJ5 B4PVB9 B4IBG9 A0A2J7PQK3 A0A182FCX3 A0A1W7R6Y5 A0A1A9ZZV3 A0A182QYC3 A0A084WCP8 A0A182VI12 A0A182YC33 A0A154NY75 W5JR02 A0A182J4M0 N6TSR9 U4U3K5 B4JTV0 A0A182RKW3 A0A182LBP1 B3P3D4 Q16QI5 A0A1S4FT36 Q7Q9I6 A0A2H1VFK8 A0A182U083 A0A2W1BN39 A0A182P8T2 A0A2A4JM43 A0A182WK19 A0A2J7PZP0 A0A0L7LAQ9 A0A212F7U9 W8AYU6
A0A194PP52 A0A0L7LB63 A0A139WPD8 A0A139WPP7 A0A139WPF3 A0A195BJQ9 F4W742 A0A151IA46 A0A151JRW6 A0A158NJC3 A0A195EYM2 A0A088APY9 A0A0L7R8H0 A0A0N0BC89 A0A3L8DRQ5 E2A3Y9 E9J1T2 K7J3G0 A0A232F8U8 A0A1J1HP71 A0A0T6AZU3 A0A0L0C0J5 B4N9U7 A0A026VYJ5 A0A1B0FNH0 A0A1I8Q177 A0A1I8Q183 A0A1I8Q151 A0A1B0BV85 A0A0P8XZR3 A0A1W4UT57 A0A1W4V5H6 B3LVQ3 A0A1W4USR1 A0A0Q9X715 W8BHF6 B4K7M7 A0A0Q9X218 A0A1B0CHP3 A0A336KKP0 A0A3B0KAQ1 A0A336M8I3 A0A0Q9WGA5 A0A0R3NK49 Q29A88 A0A0R3NGP1 A0A0R3NGB7 B4LY43 B4G2L9 A0A0R3NG76 A0A0Q9WFX9 A0A0M4ETQ6 A0A2J7PQK5 A0A2J7PQK1 A0A2J7PQP1 A0A0R1E4W5 A8JR35 A0A0R1E506 A0A0R1E3C0 Q7KSE9 Q9VEJ5 B4PVB9 B4IBG9 A0A2J7PQK3 A0A182FCX3 A0A1W7R6Y5 A0A1A9ZZV3 A0A182QYC3 A0A084WCP8 A0A182VI12 A0A182YC33 A0A154NY75 W5JR02 A0A182J4M0 N6TSR9 U4U3K5 B4JTV0 A0A182RKW3 A0A182LBP1 B3P3D4 Q16QI5 A0A1S4FT36 Q7Q9I6 A0A2H1VFK8 A0A182U083 A0A2W1BN39 A0A182P8T2 A0A2A4JM43 A0A182WK19 A0A2J7PZP0 A0A0L7LAQ9 A0A212F7U9 W8AYU6
PDB
4YH7
E-value=1.63504e-23,
Score=275
Ontologies
GO
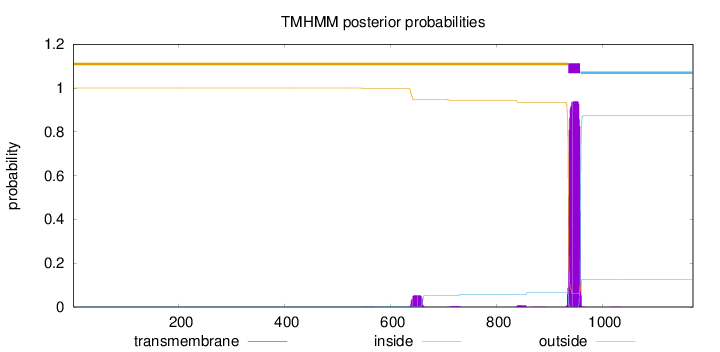
Topology
Length:
1170
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.80205
Exp number, first 60 AAs:
0.00042
Total prob of N-in:
0.00036
outside
1 - 934
TMhelix
935 - 957
inside
958 - 1170
Population Genetic Test Statistics
Pi
198.892452
Theta
152.710691
Tajima's D
0.94617
CLR
0.433555
CSRT
0.649567521623919
Interpretation
Uncertain
