Gene
KWMTBOMO03045 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005826
Annotation
PREDICTED:_15-hydroxyprostaglandin_dehydrogenase_[NAD(+)]-like?_partial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.333 Mitochondrial Reliability : 1.016 PlasmaMembrane Reliability : 1.005
Sequence
CDS
ATGAAATTACTTCTATTCCTGGTACTCGCCATCTCTGCATACCAGTGTTTTGGCTACCCAAAAAAACCAATAGATTCTCAATACGAAGTGAAAAACAAAGTAATTATTGTTACTGGAGCTGCTCGAGGTATTGGTTATGCAATAGCCGATAATTTTCTCGAAAAGGGAGCAAAGATCATAATCATACTAGACATCGATGTGCCCAAAGGACGAGACGCCGCGGCAACTTTAAATACAAAATACGGAAAGAACAGCGCGGTTTTCATCAAGTGTGACGTCACGAAAGATCTAGATGAAGTCTCCCAACTAATCCTAAGAAACTACGACGTAGATATTTTGGTAAATTGCGCAGGAGTGGTCAACGAATTTGATCCTAGAAAGACGTTGACGACAAACTCAATAGCTCTTATCGAATGGTCCCTTAAGTTCAGAGAGTACATGAGAAAAGACAATGGTGGAAAAGGCGGAACGATTATCAACATTTCTTCTATATACGGGTACACAATAGACCCGTTCTTGGTTTCGTATAAGGCGTCAAAGTTTGCTGTCTTAGGGTTTTCCAAATCACTGGGTCATGAATTGAACTACAAGATTACTGGAGTCAGGGTGCTCGTTATATGTCCCGGTGCTACAGATACAAGATTGGCAAAGAACGTTAAGGTATTCCCCGAACACAACGAGCTCTTCACAAGGCTCACAGACAAAGCACAAAAGCAGAGTGCAGATGCCGTTGGTAGGGGAGTCGTGCAAGTTTACAAGTCAGCTAGAACAGGTACGGCTTGGGACATTATTGGGGGGACACCCCCTGTTGAATCGCCTCTTGCAGCATACACTTCCATGAAAGAAATCTTGGAAAGTTAA
Protein
MKLLLFLVLAISAYQCFGYPKKPIDSQYEVKNKVIIVTGAARGIGYAIADNFLEKGAKIIIILDIDVPKGRDAAATLNTKYGKNSAVFIKCDVTKDLDEVSQLILRNYDVDILVNCAGVVNEFDPRKTLTTNSIALIEWSLKFREYMRKDNGGKGGTIINISSIYGYTIDPFLVSYKASKFAVLGFSKSLGHELNYKITGVRVLVICPGATDTRLAKNVKVFPEHNELFTRLTDKAQKQSADAVGRGVVQVYKSARTGTAWDIIGGTPPVESPLAAYTSMKEILES
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9J8I4
H9J8I5
Q1HPX6
H9J8J0
H9J8I9
A0A2H1VNC8
+ More
H9J369 A0A1L8D6N5 A0A194R949 A0A2W1BR60 A0A0H4TDA9 A0A0F6Q0W0 A0A194R584 A0A212F911 A0A0L7KQW0 A0A0L7LJP6 A0A0F6Q0X5 A0A2A4JIR0 H9JWW1 H9JWW0 A0A2W1BL60 A0A0F6Q2J9 A0A2W1BSK9 H9JYK0 A0A0H4TD16 H9JWW7 D2SNK3 H9J8I6 A0A194QJQ6 A0A194R3L5 A0A212F936 I4DKW3 A0A194R4F0 S4PIB4 U5KC20 A0A0H4TH26 H9JWW6 A0A2H1VSR5 A0A0F6Q4H7 A0A2H1VR53 H9J354 H9J355 A0A0L7K2F9 A0A2A4JJW8 H9J0A6 A0A194QQX1 A0A158NDP0 A0A194QRC6 A0A212F0G1 A0A0J7NFD4 A0A151WLU8 A0A151XEI7 B0XI43 A0A0P4WG45 A0A194Q8J5 B0XI42 A0A0J7L0X1 A0A3G1T1B2 F4W474 A0A195ET92 A0A195B6A6 A0A195BP85 A0A158NZK6 A0A194QE27 A0A194QA61 A0A023F7A3 A0A1L8D6X2 B0XCJ1 A0A026WRA5 A0A1S4G7W1 A0A076FT24 A0A139WKA8 A0A195CN33 A0A151X1L8 A0A0F6Q4J7 H9J0A4 A0A195CI97 A0A232EGH1 A0A0L7QT43 A0A0N0PDG0 A0A067R8A9 A0A1W4WBZ1 H9J0A5 E2A713 A0A195EK98 A0A0H4TG49 J9HTP4 H9J0A8 J9EB03 A0A195F4X3 A0A182GQQ8 A0A158NDN9 J9HJE4 A0A131Y3M1
H9J369 A0A1L8D6N5 A0A194R949 A0A2W1BR60 A0A0H4TDA9 A0A0F6Q0W0 A0A194R584 A0A212F911 A0A0L7KQW0 A0A0L7LJP6 A0A0F6Q0X5 A0A2A4JIR0 H9JWW1 H9JWW0 A0A2W1BL60 A0A0F6Q2J9 A0A2W1BSK9 H9JYK0 A0A0H4TD16 H9JWW7 D2SNK3 H9J8I6 A0A194QJQ6 A0A194R3L5 A0A212F936 I4DKW3 A0A194R4F0 S4PIB4 U5KC20 A0A0H4TH26 H9JWW6 A0A2H1VSR5 A0A0F6Q4H7 A0A2H1VR53 H9J354 H9J355 A0A0L7K2F9 A0A2A4JJW8 H9J0A6 A0A194QQX1 A0A158NDP0 A0A194QRC6 A0A212F0G1 A0A0J7NFD4 A0A151WLU8 A0A151XEI7 B0XI43 A0A0P4WG45 A0A194Q8J5 B0XI42 A0A0J7L0X1 A0A3G1T1B2 F4W474 A0A195ET92 A0A195B6A6 A0A195BP85 A0A158NZK6 A0A194QE27 A0A194QA61 A0A023F7A3 A0A1L8D6X2 B0XCJ1 A0A026WRA5 A0A1S4G7W1 A0A076FT24 A0A139WKA8 A0A195CN33 A0A151X1L8 A0A0F6Q4J7 H9J0A4 A0A195CI97 A0A232EGH1 A0A0L7QT43 A0A0N0PDG0 A0A067R8A9 A0A1W4WBZ1 H9J0A5 E2A713 A0A195EK98 A0A0H4TG49 J9HTP4 H9J0A8 J9EB03 A0A195F4X3 A0A182GQQ8 A0A158NDN9 J9HJE4 A0A131Y3M1
Pubmed
EMBL
BABH01027525
BABH01027529
BABH01027530
DQ443276
ABF51365.1
BABH01027538
+ More
ODYU01003471 SOQ42306.1 BABH01036649 GEYN01000013 JAV02116.1 KQ460779 KPJ12396.1 KZ149989 PZC75627.1 KP899549 AKQ06148.1 KP237884 AKD01737.1 KPJ12400.1 AGBW02009651 OWR50236.1 JTDY01007189 KOB65394.1 JTDY01000876 KOB75589.1 KP237899 AKD01752.1 NWSH01001322 PCG71699.1 BABH01043464 BABH01043465 PZC75628.1 KP237873 AKD01726.1 PZC75626.1 BABH01044234 KP899551 AKQ06150.1 BABH01040078 EZ407131 ACX53694.1 BABH01027533 KQ459144 KPJ03686.1 KPJ12398.1 OWR50237.1 AK401931 BAM18553.1 KPJ03685.1 KPJ12399.1 GAIX01005455 JAA87105.1 KC007343 AGQ45608.1 KP899550 AKQ06149.1 ODYU01003911 SOQ43254.1 KP237875 AKD01728.1 SOQ43256.1 BABH01036642 JTDY01017041 KOB51868.1 PCG71700.1 BABH01019209 KQ461175 KPJ07892.1 ADTU01012731 KPJ07894.1 AGBW02011082 OWR47236.1 LBMM01005734 KMQ91250.1 KQ982944 KYQ48859.1 KQ982254 KYQ58773.1 DS233256 EDS28983.1 GDRN01065431 JAI64685.1 KQ459302 KPJ01719.1 EDS28982.1 LBMM01001364 KMQ96442.1 MG846919 AXY94771.1 GL887491 EGI71070.1 KQ981979 KYN31470.1 KQ976587 KYM79714.1 KQ976424 KYM88327.1 ADTU01004714 KPJ01716.1 KQ459249 KPJ02362.1 GBBI01001633 JAC17079.1 GEYN01000012 JAV02117.1 DS232698 EDS44853.1 KK107119 QOIP01000009 EZA58570.1 RLU18591.1 KF960797 AII21999.1 KQ971329 KYB28353.1 KQ977600 KYN01499.1 KQ982585 KYQ54299.1 KP237895 AKD01748.1 BABH01019201 BABH01019202 KQ977720 KYN00433.1 NNAY01004765 OXU17447.1 KQ414755 KOC61724.1 KQ460366 KPJ15602.1 KK852815 KDR15798.1 BABH01019208 GL437252 EFN70745.1 KQ978747 KYN28685.1 KP899548 AKQ06147.1 CH477964 EJY58013.1 BABH01019210 CH477766 EJY57916.1 KQ981820 KYN35129.1 JXUM01081117 KQ563228 KXJ74247.1 EJY58014.1 GEFM01002721 JAP73075.1
ODYU01003471 SOQ42306.1 BABH01036649 GEYN01000013 JAV02116.1 KQ460779 KPJ12396.1 KZ149989 PZC75627.1 KP899549 AKQ06148.1 KP237884 AKD01737.1 KPJ12400.1 AGBW02009651 OWR50236.1 JTDY01007189 KOB65394.1 JTDY01000876 KOB75589.1 KP237899 AKD01752.1 NWSH01001322 PCG71699.1 BABH01043464 BABH01043465 PZC75628.1 KP237873 AKD01726.1 PZC75626.1 BABH01044234 KP899551 AKQ06150.1 BABH01040078 EZ407131 ACX53694.1 BABH01027533 KQ459144 KPJ03686.1 KPJ12398.1 OWR50237.1 AK401931 BAM18553.1 KPJ03685.1 KPJ12399.1 GAIX01005455 JAA87105.1 KC007343 AGQ45608.1 KP899550 AKQ06149.1 ODYU01003911 SOQ43254.1 KP237875 AKD01728.1 SOQ43256.1 BABH01036642 JTDY01017041 KOB51868.1 PCG71700.1 BABH01019209 KQ461175 KPJ07892.1 ADTU01012731 KPJ07894.1 AGBW02011082 OWR47236.1 LBMM01005734 KMQ91250.1 KQ982944 KYQ48859.1 KQ982254 KYQ58773.1 DS233256 EDS28983.1 GDRN01065431 JAI64685.1 KQ459302 KPJ01719.1 EDS28982.1 LBMM01001364 KMQ96442.1 MG846919 AXY94771.1 GL887491 EGI71070.1 KQ981979 KYN31470.1 KQ976587 KYM79714.1 KQ976424 KYM88327.1 ADTU01004714 KPJ01716.1 KQ459249 KPJ02362.1 GBBI01001633 JAC17079.1 GEYN01000012 JAV02117.1 DS232698 EDS44853.1 KK107119 QOIP01000009 EZA58570.1 RLU18591.1 KF960797 AII21999.1 KQ971329 KYB28353.1 KQ977600 KYN01499.1 KQ982585 KYQ54299.1 KP237895 AKD01748.1 BABH01019201 BABH01019202 KQ977720 KYN00433.1 NNAY01004765 OXU17447.1 KQ414755 KOC61724.1 KQ460366 KPJ15602.1 KK852815 KDR15798.1 BABH01019208 GL437252 EFN70745.1 KQ978747 KYN28685.1 KP899548 AKQ06147.1 CH477964 EJY58013.1 BABH01019210 CH477766 EJY57916.1 KQ981820 KYN35129.1 JXUM01081117 KQ563228 KXJ74247.1 EJY58014.1 GEFM01002721 JAP73075.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000037510
UP000218220
UP000053268
+ More
UP000005205 UP000036403 UP000075809 UP000002320 UP000007755 UP000078541 UP000078540 UP000053097 UP000279307 UP000007266 UP000078542 UP000215335 UP000053825 UP000027135 UP000192223 UP000000311 UP000078492 UP000008820 UP000069940 UP000249989
UP000005205 UP000036403 UP000075809 UP000002320 UP000007755 UP000078541 UP000078540 UP000053097 UP000279307 UP000007266 UP000078542 UP000215335 UP000053825 UP000027135 UP000192223 UP000000311 UP000078492 UP000008820 UP000069940 UP000249989
PRIDE
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9J8I4
H9J8I5
Q1HPX6
H9J8J0
H9J8I9
A0A2H1VNC8
+ More
H9J369 A0A1L8D6N5 A0A194R949 A0A2W1BR60 A0A0H4TDA9 A0A0F6Q0W0 A0A194R584 A0A212F911 A0A0L7KQW0 A0A0L7LJP6 A0A0F6Q0X5 A0A2A4JIR0 H9JWW1 H9JWW0 A0A2W1BL60 A0A0F6Q2J9 A0A2W1BSK9 H9JYK0 A0A0H4TD16 H9JWW7 D2SNK3 H9J8I6 A0A194QJQ6 A0A194R3L5 A0A212F936 I4DKW3 A0A194R4F0 S4PIB4 U5KC20 A0A0H4TH26 H9JWW6 A0A2H1VSR5 A0A0F6Q4H7 A0A2H1VR53 H9J354 H9J355 A0A0L7K2F9 A0A2A4JJW8 H9J0A6 A0A194QQX1 A0A158NDP0 A0A194QRC6 A0A212F0G1 A0A0J7NFD4 A0A151WLU8 A0A151XEI7 B0XI43 A0A0P4WG45 A0A194Q8J5 B0XI42 A0A0J7L0X1 A0A3G1T1B2 F4W474 A0A195ET92 A0A195B6A6 A0A195BP85 A0A158NZK6 A0A194QE27 A0A194QA61 A0A023F7A3 A0A1L8D6X2 B0XCJ1 A0A026WRA5 A0A1S4G7W1 A0A076FT24 A0A139WKA8 A0A195CN33 A0A151X1L8 A0A0F6Q4J7 H9J0A4 A0A195CI97 A0A232EGH1 A0A0L7QT43 A0A0N0PDG0 A0A067R8A9 A0A1W4WBZ1 H9J0A5 E2A713 A0A195EK98 A0A0H4TG49 J9HTP4 H9J0A8 J9EB03 A0A195F4X3 A0A182GQQ8 A0A158NDN9 J9HJE4 A0A131Y3M1
H9J369 A0A1L8D6N5 A0A194R949 A0A2W1BR60 A0A0H4TDA9 A0A0F6Q0W0 A0A194R584 A0A212F911 A0A0L7KQW0 A0A0L7LJP6 A0A0F6Q0X5 A0A2A4JIR0 H9JWW1 H9JWW0 A0A2W1BL60 A0A0F6Q2J9 A0A2W1BSK9 H9JYK0 A0A0H4TD16 H9JWW7 D2SNK3 H9J8I6 A0A194QJQ6 A0A194R3L5 A0A212F936 I4DKW3 A0A194R4F0 S4PIB4 U5KC20 A0A0H4TH26 H9JWW6 A0A2H1VSR5 A0A0F6Q4H7 A0A2H1VR53 H9J354 H9J355 A0A0L7K2F9 A0A2A4JJW8 H9J0A6 A0A194QQX1 A0A158NDP0 A0A194QRC6 A0A212F0G1 A0A0J7NFD4 A0A151WLU8 A0A151XEI7 B0XI43 A0A0P4WG45 A0A194Q8J5 B0XI42 A0A0J7L0X1 A0A3G1T1B2 F4W474 A0A195ET92 A0A195B6A6 A0A195BP85 A0A158NZK6 A0A194QE27 A0A194QA61 A0A023F7A3 A0A1L8D6X2 B0XCJ1 A0A026WRA5 A0A1S4G7W1 A0A076FT24 A0A139WKA8 A0A195CN33 A0A151X1L8 A0A0F6Q4J7 H9J0A4 A0A195CI97 A0A232EGH1 A0A0L7QT43 A0A0N0PDG0 A0A067R8A9 A0A1W4WBZ1 H9J0A5 E2A713 A0A195EK98 A0A0H4TG49 J9HTP4 H9J0A8 J9EB03 A0A195F4X3 A0A182GQQ8 A0A158NDN9 J9HJE4 A0A131Y3M1
PDB
5ILO
E-value=2.17238e-29,
Score=320
Ontologies
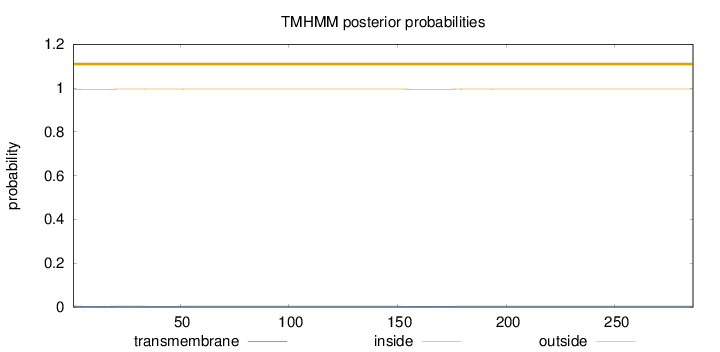
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.998740
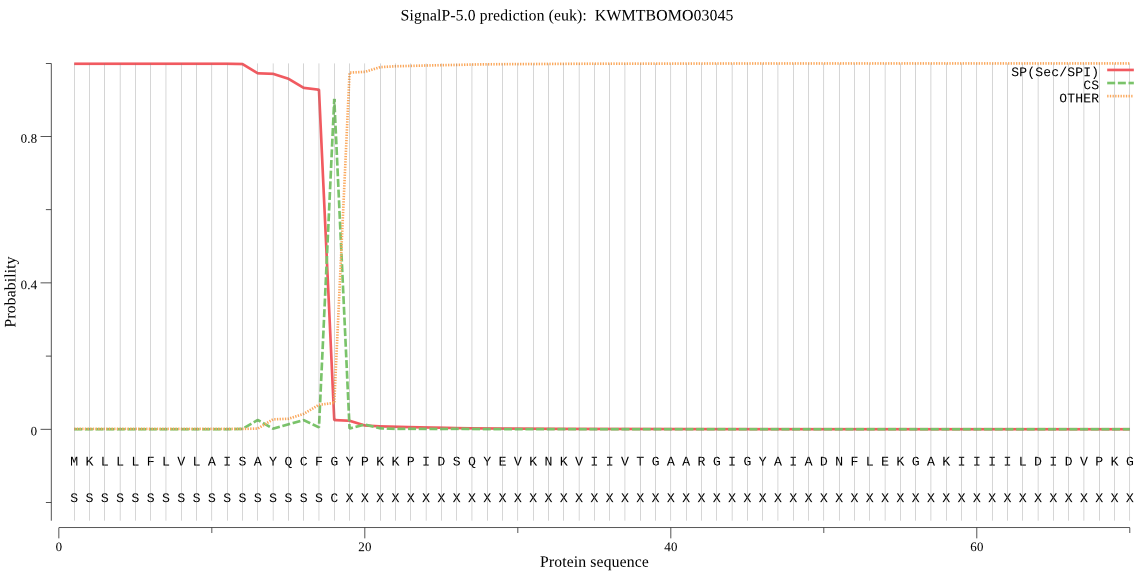
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15986
Exp number, first 60 AAs:
0.06426
Total prob of N-in:
0.00570
outside
1 - 286
Population Genetic Test Statistics
Pi
309.765087
Theta
166.96763
Tajima's D
2.806327
CLR
0.290784
CSRT
0.968451577421129
Interpretation
Possibly Balancing Selection
