Gene
KWMTBOMO03037 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005831
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_15-hydroxyprostaglandin_dehydrogenase_[NAD(+)]-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.872
Sequence
CDS
ATGTACCAAAAGGTAGAAGGTAGACAAGCTGCGAAAGAGTTAAATTCTAAATATGGAAAGAATAAAGCTGAGTTCATTGAATGCGATATTACTAAAGACTTGGAGAGGGTCTCGAAAATGATTTATAAGAAATACAAGTATGTGGATGTATTGGTTAACAATGCAGAAATAGGTGAAGAAAATAAACCAAAGAAACTCCTGCTGACAAATGTTGTAGCTACTATTGAATTTTCTCTCAAGTTCATGGAAAATATGAGAAAAGACAAACTAGGCGGAAAAGGAGGTACTATAATCAATATAGCGTCAATAGTAGTGCATTACATTGATTCTTTCTTCTTTACGTACAGAGGAACGAAATTTGCCATTATGGGATTCTCACGATCTTTAGGACACGAATGCAACTATAAAAAGAGCGGTTTGAGAGTACTAACAATATGTCCGGGAGCAACAAATACAACACTGTGTTCAGATAATCCCAAAATGAATAGTGAAGACGCGGCAGCTCTTGCTAAAGAATATCAATATCAAATTATACAATCTCCCGACAATGTAGGGAAAGGTGCTGTGGAAATATTCAAAAATGCTAATACTGGTACGTCTTGGGAAGTCGCGTATGAAAAATATCCTGTTGAGTCGACTGATACGATAAGAGTTTAA
Protein
MYQKVEGRQAAKELNSKYGKNKAEFIECDITKDLERVSKMIYKKYKYVDVLVNNAEIGEENKPKKLLLTNVVATIEFSLKFMENMRKDKLGGKGGTIINIASIVVHYIDSFFFTYRGTKFAIMGFSRSLGHECNYKKSGLRVLTICPGATNTTLCSDNPKMNSEDAAALAKEYQYQIIQSPDNVGKGAVEIFKNANTGTSWEVAYEKYPVESTDTIRV
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9J8I9
Q1HPX6
H9J8J0
H9J8I8
H9J8I6
H9J8I4
+ More
A0A0L7KQW0 H9J8I5 A0A2A4JIR0 S4PIB4 D2SNK3 A0A194R4F0 A0A0H4TDA9 A0A2W1BSK9 A0A0L7LJP6 A0A0F6Q0X5 U5KC20 A0A194QJQ6 A0A212F911 A0A194R949 A0A0H4TD16 I4DKW3 A0A2H1VNC8 A0A1L8D6N5 A0A194R584 A0A2W1BR60 A0A0F6Q2J9 A0A0F6Q0W0 A0A194R3L5 H9JWW1 H9J369 A0A2H1VSR5 H9JWW7 A0A0F6Q4H7 H9JWW0 H9JYK0 A0A2W1BL60 A0A0L7K392 A0A076FT29 A0A212F936 A0A0H4TH26 H9JWW6 H9J354 A0A2H1VR53 A0A2A4JJW8 A0A0L7K2F9 H9J355 A0A194QQX1 A0A194Q8J5 H9J0A8 A0A194QRC6 A0A194QE27 A0A3G1T1B2 H9J0A6 H9J353 A0A076FT24 H9J0A5 A0A212F0G1 H9J0A4 A0A0L7KNH1 Q6RUR3 A0A158NDP0 A0A195BP85 A0A0H4TG49 A0A0F6Q4J7 A0A224XI78 A0A0J7NFD4 A0A2W1BM47 A0A023F7A3 A0A0H4TY65 A0A139WKA8 A0A1Y1LZE8 R4FLI9 A0A212F0F9 A0A1Y1K7W8 A0A224XI56 E2A712 A0A151XEI7 A0A1L8D6X2 A0A2A4K9E1 A0A182GQQ7 A0A1Y1MVB3 A0A140KPR8 A0A0J7L0X1 J9HJ88 A0A2A4KAR2 B0XCJ3 D6WGE4 A0A182H9W1 D6WGE5 J3JY86 A0A151WLU8 A0A2W1BJY5 B0XI43 E2BHN0
A0A0L7KQW0 H9J8I5 A0A2A4JIR0 S4PIB4 D2SNK3 A0A194R4F0 A0A0H4TDA9 A0A2W1BSK9 A0A0L7LJP6 A0A0F6Q0X5 U5KC20 A0A194QJQ6 A0A212F911 A0A194R949 A0A0H4TD16 I4DKW3 A0A2H1VNC8 A0A1L8D6N5 A0A194R584 A0A2W1BR60 A0A0F6Q2J9 A0A0F6Q0W0 A0A194R3L5 H9JWW1 H9J369 A0A2H1VSR5 H9JWW7 A0A0F6Q4H7 H9JWW0 H9JYK0 A0A2W1BL60 A0A0L7K392 A0A076FT29 A0A212F936 A0A0H4TH26 H9JWW6 H9J354 A0A2H1VR53 A0A2A4JJW8 A0A0L7K2F9 H9J355 A0A194QQX1 A0A194Q8J5 H9J0A8 A0A194QRC6 A0A194QE27 A0A3G1T1B2 H9J0A6 H9J353 A0A076FT24 H9J0A5 A0A212F0G1 H9J0A4 A0A0L7KNH1 Q6RUR3 A0A158NDP0 A0A195BP85 A0A0H4TG49 A0A0F6Q4J7 A0A224XI78 A0A0J7NFD4 A0A2W1BM47 A0A023F7A3 A0A0H4TY65 A0A139WKA8 A0A1Y1LZE8 R4FLI9 A0A212F0F9 A0A1Y1K7W8 A0A224XI56 E2A712 A0A151XEI7 A0A1L8D6X2 A0A2A4K9E1 A0A182GQQ7 A0A1Y1MVB3 A0A140KPR8 A0A0J7L0X1 J9HJ88 A0A2A4KAR2 B0XCJ3 D6WGE4 A0A182H9W1 D6WGE5 J3JY86 A0A151WLU8 A0A2W1BJY5 B0XI43 E2BHN0
Pubmed
EMBL
BABH01027538
DQ443276
ABF51365.1
BABH01027535
BABH01027536
BABH01027537
+ More
BABH01027533 BABH01027525 JTDY01007189 KOB65394.1 BABH01027529 BABH01027530 NWSH01001322 PCG71699.1 GAIX01005455 JAA87105.1 EZ407131 ACX53694.1 KQ460779 KPJ12399.1 KP899549 AKQ06148.1 KZ149989 PZC75626.1 JTDY01000876 KOB75589.1 KP237899 AKD01752.1 KC007343 AGQ45608.1 KQ459144 KPJ03686.1 AGBW02009651 OWR50236.1 KPJ12396.1 KP899551 AKQ06150.1 AK401931 BAM18553.1 KPJ03685.1 ODYU01003471 SOQ42306.1 GEYN01000013 JAV02116.1 KPJ12400.1 PZC75627.1 KP237873 AKD01726.1 KP237884 AKD01737.1 KPJ12398.1 BABH01043464 BABH01036649 ODYU01003911 SOQ43254.1 BABH01040078 KP237875 AKD01728.1 BABH01043465 BABH01044234 PZC75628.1 JTDY01013396 KOB52144.1 KF960802 AII22004.1 OWR50237.1 KP899550 AKQ06149.1 BABH01036642 SOQ43256.1 PCG71700.1 JTDY01017041 KOB51868.1 KQ461175 KPJ07892.1 KQ459302 KPJ01719.1 BABH01019210 KPJ07894.1 KPJ01716.1 MG846919 AXY94771.1 BABH01019209 BABH01036648 KF960797 AII21999.1 BABH01019208 AGBW02011082 OWR47236.1 BABH01019201 BABH01019202 JTDY01007968 KOB64832.1 AY491503 AAR84629.1 ADTU01012731 KQ976424 KYM88327.1 KP899548 AKQ06147.1 KP237895 AKD01748.1 GFTR01004241 JAW12185.1 LBMM01005734 KMQ91250.1 KZ149998 PZC75361.1 GBBI01001633 JAC17079.1 KP899552 AKQ06151.1 KQ971329 KYB28353.1 GEZM01045515 JAV77770.1 ACPB03004191 GAHY01001368 JAA76142.1 OWR47238.1 GEZM01089353 JAV57529.1 GFTR01004259 JAW12167.1 GL437252 EFN70744.1 KQ982254 KYQ58773.1 GEYN01000012 JAV02117.1 NWSH01000014 PCG80831.1 JXUM01081116 KQ563228 KXJ74246.1 GEZM01023447 JAV88385.1 LN794625 CEO43692.1 LBMM01001364 KMQ96442.1 CH477766 EJY57919.1 PCG80830.1 DS232698 EDS44855.1 EFA01118.2 JXUM01030715 JXUM01030716 JXUM01030717 KQ560893 KXJ80459.1 EFA01117.2 BT128210 AEE63171.1 KQ982944 KYQ48859.1 PZC75359.1 DS233256 EDS28983.1 GL448322 EFN84778.1
BABH01027533 BABH01027525 JTDY01007189 KOB65394.1 BABH01027529 BABH01027530 NWSH01001322 PCG71699.1 GAIX01005455 JAA87105.1 EZ407131 ACX53694.1 KQ460779 KPJ12399.1 KP899549 AKQ06148.1 KZ149989 PZC75626.1 JTDY01000876 KOB75589.1 KP237899 AKD01752.1 KC007343 AGQ45608.1 KQ459144 KPJ03686.1 AGBW02009651 OWR50236.1 KPJ12396.1 KP899551 AKQ06150.1 AK401931 BAM18553.1 KPJ03685.1 ODYU01003471 SOQ42306.1 GEYN01000013 JAV02116.1 KPJ12400.1 PZC75627.1 KP237873 AKD01726.1 KP237884 AKD01737.1 KPJ12398.1 BABH01043464 BABH01036649 ODYU01003911 SOQ43254.1 BABH01040078 KP237875 AKD01728.1 BABH01043465 BABH01044234 PZC75628.1 JTDY01013396 KOB52144.1 KF960802 AII22004.1 OWR50237.1 KP899550 AKQ06149.1 BABH01036642 SOQ43256.1 PCG71700.1 JTDY01017041 KOB51868.1 KQ461175 KPJ07892.1 KQ459302 KPJ01719.1 BABH01019210 KPJ07894.1 KPJ01716.1 MG846919 AXY94771.1 BABH01019209 BABH01036648 KF960797 AII21999.1 BABH01019208 AGBW02011082 OWR47236.1 BABH01019201 BABH01019202 JTDY01007968 KOB64832.1 AY491503 AAR84629.1 ADTU01012731 KQ976424 KYM88327.1 KP899548 AKQ06147.1 KP237895 AKD01748.1 GFTR01004241 JAW12185.1 LBMM01005734 KMQ91250.1 KZ149998 PZC75361.1 GBBI01001633 JAC17079.1 KP899552 AKQ06151.1 KQ971329 KYB28353.1 GEZM01045515 JAV77770.1 ACPB03004191 GAHY01001368 JAA76142.1 OWR47238.1 GEZM01089353 JAV57529.1 GFTR01004259 JAW12167.1 GL437252 EFN70744.1 KQ982254 KYQ58773.1 GEYN01000012 JAV02117.1 NWSH01000014 PCG80831.1 JXUM01081116 KQ563228 KXJ74246.1 GEZM01023447 JAV88385.1 LN794625 CEO43692.1 LBMM01001364 KMQ96442.1 CH477766 EJY57919.1 PCG80830.1 DS232698 EDS44855.1 EFA01118.2 JXUM01030715 JXUM01030716 JXUM01030717 KQ560893 KXJ80459.1 EFA01117.2 BT128210 AEE63171.1 KQ982944 KYQ48859.1 PZC75359.1 DS233256 EDS28983.1 GL448322 EFN84778.1
Proteomes
PRIDE
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9J8I9
Q1HPX6
H9J8J0
H9J8I8
H9J8I6
H9J8I4
+ More
A0A0L7KQW0 H9J8I5 A0A2A4JIR0 S4PIB4 D2SNK3 A0A194R4F0 A0A0H4TDA9 A0A2W1BSK9 A0A0L7LJP6 A0A0F6Q0X5 U5KC20 A0A194QJQ6 A0A212F911 A0A194R949 A0A0H4TD16 I4DKW3 A0A2H1VNC8 A0A1L8D6N5 A0A194R584 A0A2W1BR60 A0A0F6Q2J9 A0A0F6Q0W0 A0A194R3L5 H9JWW1 H9J369 A0A2H1VSR5 H9JWW7 A0A0F6Q4H7 H9JWW0 H9JYK0 A0A2W1BL60 A0A0L7K392 A0A076FT29 A0A212F936 A0A0H4TH26 H9JWW6 H9J354 A0A2H1VR53 A0A2A4JJW8 A0A0L7K2F9 H9J355 A0A194QQX1 A0A194Q8J5 H9J0A8 A0A194QRC6 A0A194QE27 A0A3G1T1B2 H9J0A6 H9J353 A0A076FT24 H9J0A5 A0A212F0G1 H9J0A4 A0A0L7KNH1 Q6RUR3 A0A158NDP0 A0A195BP85 A0A0H4TG49 A0A0F6Q4J7 A0A224XI78 A0A0J7NFD4 A0A2W1BM47 A0A023F7A3 A0A0H4TY65 A0A139WKA8 A0A1Y1LZE8 R4FLI9 A0A212F0F9 A0A1Y1K7W8 A0A224XI56 E2A712 A0A151XEI7 A0A1L8D6X2 A0A2A4K9E1 A0A182GQQ7 A0A1Y1MVB3 A0A140KPR8 A0A0J7L0X1 J9HJ88 A0A2A4KAR2 B0XCJ3 D6WGE4 A0A182H9W1 D6WGE5 J3JY86 A0A151WLU8 A0A2W1BJY5 B0XI43 E2BHN0
A0A0L7KQW0 H9J8I5 A0A2A4JIR0 S4PIB4 D2SNK3 A0A194R4F0 A0A0H4TDA9 A0A2W1BSK9 A0A0L7LJP6 A0A0F6Q0X5 U5KC20 A0A194QJQ6 A0A212F911 A0A194R949 A0A0H4TD16 I4DKW3 A0A2H1VNC8 A0A1L8D6N5 A0A194R584 A0A2W1BR60 A0A0F6Q2J9 A0A0F6Q0W0 A0A194R3L5 H9JWW1 H9J369 A0A2H1VSR5 H9JWW7 A0A0F6Q4H7 H9JWW0 H9JYK0 A0A2W1BL60 A0A0L7K392 A0A076FT29 A0A212F936 A0A0H4TH26 H9JWW6 H9J354 A0A2H1VR53 A0A2A4JJW8 A0A0L7K2F9 H9J355 A0A194QQX1 A0A194Q8J5 H9J0A8 A0A194QRC6 A0A194QE27 A0A3G1T1B2 H9J0A6 H9J353 A0A076FT24 H9J0A5 A0A212F0G1 H9J0A4 A0A0L7KNH1 Q6RUR3 A0A158NDP0 A0A195BP85 A0A0H4TG49 A0A0F6Q4J7 A0A224XI78 A0A0J7NFD4 A0A2W1BM47 A0A023F7A3 A0A0H4TY65 A0A139WKA8 A0A1Y1LZE8 R4FLI9 A0A212F0F9 A0A1Y1K7W8 A0A224XI56 E2A712 A0A151XEI7 A0A1L8D6X2 A0A2A4K9E1 A0A182GQQ7 A0A1Y1MVB3 A0A140KPR8 A0A0J7L0X1 J9HJ88 A0A2A4KAR2 B0XCJ3 D6WGE4 A0A182H9W1 D6WGE5 J3JY86 A0A151WLU8 A0A2W1BJY5 B0XI43 E2BHN0
PDB
2GDZ
E-value=2.63008e-14,
Score=188
Ontologies
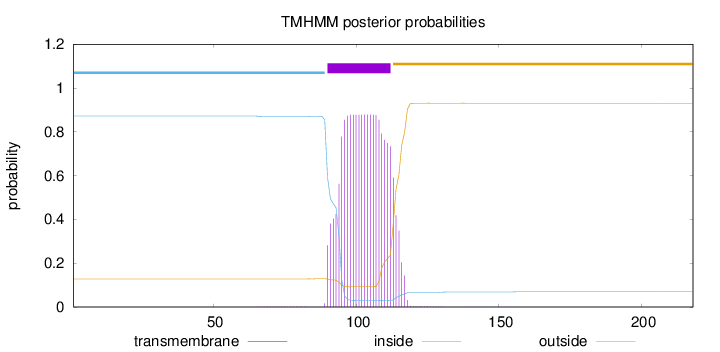
Topology
Length:
218
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.04377
Exp number, first 60 AAs:
0
Total prob of N-in:
0.87222
inside
1 - 89
TMhelix
90 - 112
outside
113 - 218
Population Genetic Test Statistics
Pi
501.259678
Theta
223.768647
Tajima's D
3.923693
CLR
0.730527
CSRT
0.998450077496125
Interpretation
Uncertain
