Gene
KWMTBOMO03031
Annotation
PREDICTED:_uncharacterized_protein_LOC107445205_[Parasteatoda_tepidariorum]
Location in the cell
Nuclear Reliability : 4.008
Sequence
CDS
ATGGTATATAAATACAAGAGGAAGACTAATCAAGCCCAATGGAGTGAGGACAGTATGATGCTTGCAATAGCAGATTGCAATTCTGGCATTCCTGTCAAAACGACCGCAAAAAAGTATGGTCTACCTTACGCCACTCTCTATAGGCATTGGAAAAAAGGTAGTTCGACGGCTCAGCTTGGTCGTTTTCGTAAAGTTTTCAACGATGGTCAAGAGGGAGATCTTCGAACGTATTTGTATGAAATGGATAGTGTTTTTTATGGGCTAACCCGAGAAGATTTTAAAACATTAGTTTTTGAATACGCGAAACGCAATAATGTCACTTATCCGCCTAGTTGGGACAAAAATAAAAAAGCTGGTGATGATTGGCTAGCAGGATTTATTAGAAGAAATCCACAAATTACACTAAGAATTCCAGAAGCAACCACAATTGGCAGAGTTAAAGGTTTCAGTCGTCCTCAAGTAGAAAGGTTCTATCTATGTCAAATTGATGGTACTAGAATCTATAATATAGATGAAACTGGCATTCAGACCTCGACAAATAAACCACCCAAGGTTCTTTCAGTTAAAGCTTTACAGTACGCAACTGAAAATAATGTCATCATTTTGTCTTTGCCACCGCACACCTCAAACAAACTTCAACCTCTGGACGTGGCTGTGTATGGTCCATTTAAAATATTCTTCGAGCAAGAAATCAATAGATTTCAAAAGGCTCATCCAGGTAGGATAATCAACCAATACGATGTAGCCAAACTGTTCAGTCCAGCTTATCTGAAATCTGCTACTCCTAACAATGCTATCAAAGGCTTTCAATCAACCGGTATCTGGCCAACAAATAAAGATATATGGGGAGAAGAAGATTATGTACCATCTTCTATTACAATGACAGACCGTAATACCGCTAACCAACTGGAGACTTTTATAGACTCAAACAGTCAAAATGTTGAAATTACGATTGACATCGAAAAGCCTTCTACATCAGGGCTCCAAAGTCAGTCACTGCAAATTGATGTAGAACCACTGGAACAGATTGAAAGAAAATTTATTGAGAATGTGAATGTAGAAGTTGAAGCTGCAGTGGAAAATAGTATCATGGGTATAGTATCAGTGTCCAATATGAATCAAATACATTCTCGAGATACATCGCCTTCAATTTTGGATAAATTACCCGACAAAGATCCCCTTACATGCAATGCTGTTGTAGTAACGGCTGTAGCTGATAATACTCAAAAATTGTCTAAACCACAAGAAGCAATGTCCACTGAAGATATGAATCAAGGTGCAGATCTGACTCAAGACAGTATAGATATAAAAGAGGGTCAATTTAAAGGCAACGATGATGAAATTATTATGGACCTGGAATCATCTACGGCTCGGGCTTCAGCATATTTCAGTCCTAAAGACATAAGACCGCTTCCGATGCCACGCGTTTCTGTGGGTACACGTAAAAGAAAGGTTCAAAAGTCTGAGGTGTTAACAAGCACGCCAGTGAAGCGAAAACAAAAAGAAAAATTTATAAAAATTAATGTCAAAGCAATGAAAAATATGAATATAGATTTAAATGCTAAGCCAAAGACTGCGACAAAGAAAAATACTAAAACGACCAAAAATATAATTGTTAAAGATAAAAGTAGTGATAAGGAAAACGAGGAAGAACAGTGTGTATGCTATTATTGTGGGGAGAACTATATAGAAATAAATAAAAAACCAGTTGAGGATTGGATTCAGTGTGATAAATGTAAGCAACGGAGTCACGAAAAATGCACGGCTTATGGAGGAATAGGCTTATTCTTTTGTGATATTTGCACGGATTAG
Protein
MVYKYKRKTNQAQWSEDSMMLAIADCNSGIPVKTTAKKYGLPYATLYRHWKKGSSTAQLGRFRKVFNDGQEGDLRTYLYEMDSVFYGLTREDFKTLVFEYAKRNNVTYPPSWDKNKKAGDDWLAGFIRRNPQITLRIPEATTIGRVKGFSRPQVERFYLCQIDGTRIYNIDETGIQTSTNKPPKVLSVKALQYATENNVIILSLPPHTSNKLQPLDVAVYGPFKIFFEQEINRFQKAHPGRIINQYDVAKLFSPAYLKSATPNNAIKGFQSTGIWPTNKDIWGEEDYVPSSITMTDRNTANQLETFIDSNSQNVEITIDIEKPSTSGLQSQSLQIDVEPLEQIERKFIENVNVEVEAAVENSIMGIVSVSNMNQIHSRDTSPSILDKLPDKDPLTCNAVVVTAVADNTQKLSKPQEAMSTEDMNQGADLTQDSIDIKEGQFKGNDDEIIMDLESSTARASAYFSPKDIRPLPMPRVSVGTRKRKVQKSEVLTSTPVKRKQKEKFIKINVKAMKNMNIDLNAKPKTATKKNTKTTKNIIVKDKSSDKENEEEQCVCYYCGENYIEINKKPVEDWIQCDKCKQRSHEKCTAYGGIGLFFCDICTD
Summary
Uniprot
Pubmed
EMBL
Proteomes
Pfam
PF03184 DDE_1
Interpro
SUPFAM
SSF57903
SSF57903
Gene 3D
ProteinModelPortal
Ontologies
KEGG
GO
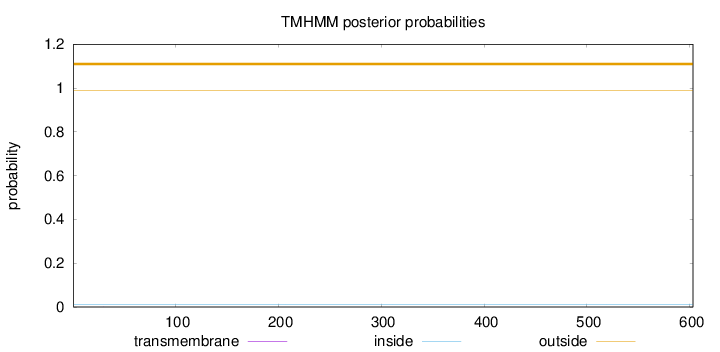
Topology
Length:
603
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00419999999999999
Exp number, first 60 AAs:
0.00241
Total prob of N-in:
0.01246
outside
1 - 603
Population Genetic Test Statistics
Pi
463.568802
Theta
203.939837
Tajima's D
4.054084
CLR
0
CSRT
0.999150042497875
Interpretation
Uncertain
