Gene
KWMTBOMO03030 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005742
Annotation
PREDICTED:_atlastin-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.638
Sequence
CDS
ATGAGCAGTCTCGGTGAAGCCAAAGGGGTGGAGGTGGTGAAGGTGACCGACGACCACACGTATCAGCTGAATGAGGATGCATTACGGATGATCCTACTGCGAGATGAGGTCAAGGACCTTCCTGTGGTCGTAGTCAGCGTCGCTGGCGTCTACCGCGGTGGAAAGTCCTTCTTGCTGGACTTCTTCCTCAGATACCTAAAAGCGCCCCCCTCGTCTCGTGATAGTGCTGCGTGGCTGGGCCAGGAAGACGAGCCGCTGACGGGATTCATTTGGCGGGCTGGTTGCGACCGTCAGACAACTGGTATCGTTCTATGGTCGGAACCGATTGTTACCAGATTCAACGATCAAAAGATCACTAACAATCAGCACGCGGAGTTGAGGGAATTACGAGAGCGAATCCGTTCTTGTTTCAAATCGGTCTCCTGTTTCCTGATGCCTCACCCGGGGTTCGTGGTTTCGGAGCAAAACTTTAATGGACAACTAGCTGACATTCGTATTGAGTTCAAAGAAGCGCTTCGAGATCTGGTACCATCGATGTTCGGATCTAGGAATTTGGTGCCGAAAAAAATCAATGGTCACCAAATCAAAACTCGGGACCTGTTTGATTTCTTTAAGACTTACGTGAACATCTACAACAGCGAGGAGCTGCCTACTCCGGTTACCATTTTGAAGGCAACATCTGAGGTGGCATTGATATCTGCGATCAGGGATGCAAGGGAACAGTATGAAAAACGTATGGAAATGAATGCCGGTGCCAAACAGCCGAGTGTACCAGACAACGTGCTTGAAGATCAACACAAGAGAACCGTTCTTATTATTCGTCAATCGTTTGAGAGTAAAAAGAGGATTGGTTCACAAAAGGACGCCAAGGAGCATATCGAAAAGCTGATTACGGAATTAGAAGCACGCTTGCAGCATTACTTGACATTGAACAAAGCGAAACTTCAGAAAGCCGTGGTTGACGCGAAACAAGCTTACGATGACGCAGTTCAGAAGGTGACTAAGGGAGAGCCTCTCTGTCTGCACCCCTTAGACTTAGATTCACTCCACAACAAGGCGGTGGACGCGGCGGCAGATTTATTCGATAACAACAGAAGAACTCCCGATAAGGAAAGCGACCCTGAAAGGATTTCGTTAATGAAGCATCTCGAAGGTAATATCAAAGATTTGCGATTGAAAAACGATTATAACAATAAAGTCTTCATTAGCGAGGCCCGAAAGGTATATGAGACATTAATGCAGAAGCACGTCTGGATGGGATCGTGTGTCTCTGACGAATTATTGCAGAATTGCCACAGAAATGCATTGAAGAAGGCCATTGGAATCTTGAAATCGCATCGCAACATCCCCACAAACCATAGCGAAGATAAATACATCAGTTCTTTACAAGAGAGCATCGTAGAAGAATTCCAGAAGTTTCGTTCAGCGAACAACAACGCCAATAAGATTGCGATCCAAGAGGCAGTATACTCTTACAACAATCACATGACTTCAGCCTGGGGACCGCCTTCCAGCTGCTTCCATCCCCAAGACCTAACGAAAATCCACGAGACTAATAAAGCATTGGCCCTGACTGAATTCCATAACAGCAGAAATACGACGGCTGATGATGATGGTGATGACGATGATGACGGTGATGTCAATAAACAAAAACTCGTTGAGCTATTGAACAGACGTTATAGTGAACTGAAAGAAATCAACTCGGCTTCCAACGATATGGCAGTTGCCGATTCATACCGCGAATACTGCAGGTATATGGACGGGAAATGTAAGCCGTCGGTCCTGTCTATATTAATTGTTCCTTGGCTGGTCAAGGTGATCAGACGTCTGCCAGATTATCATAAAGAAGGCAAAAACGCAGCATGGGATTACTTCAGAAGCAAGAGACGCAATTACTCCTACCGTTCCGATGACAGTTACAGAACAGACCTCGAATCTAAACTGGAAGATGGTTTTCATGAGTATTGTCACCCGCTGAACGCACTCACTCGTGAGTTCGGTATATGA
Protein
MSSLGEAKGVEVVKVTDDHTYQLNEDALRMILLRDEVKDLPVVVVSVAGVYRGGKSFLLDFFLRYLKAPPSSRDSAAWLGQEDEPLTGFIWRAGCDRQTTGIVLWSEPIVTRFNDQKITNNQHAELRELRERIRSCFKSVSCFLMPHPGFVVSEQNFNGQLADIRIEFKEALRDLVPSMFGSRNLVPKKINGHQIKTRDLFDFFKTYVNIYNSEELPTPVTILKATSEVALISAIRDAREQYEKRMEMNAGAKQPSVPDNVLEDQHKRTVLIIRQSFESKKRIGSQKDAKEHIEKLITELEARLQHYLTLNKAKLQKAVVDAKQAYDDAVQKVTKGEPLCLHPLDLDSLHNKAVDAAADLFDNNRRTPDKESDPERISLMKHLEGNIKDLRLKNDYNNKVFISEARKVYETLMQKHVWMGSCVSDELLQNCHRNALKKAIGILKSHRNIPTNHSEDKYISSLQESIVEEFQKFRSANNNANKIAIQEAVYSYNNHMTSAWGPPSSCFHPQDLTKIHETNKALALTEFHNSRNTTADDDGDDDDDGDVNKQKLVELLNRRYSELKEINSASNDMAVADSYREYCRYMDGKCKPSVLSILIVPWLVKVIRRLPDYHKEGKNAAWDYFRSKRRNYSYRSDDSYRTDLESKLEDGFHEYCHPLNALTREFGI
Summary
Uniprot
EMBL
Proteomes
Pfam
PF02263 GBP
Interpro
ProteinModelPortal
PDB
3X1D
E-value=2.63425e-36,
Score=383
Ontologies
GO
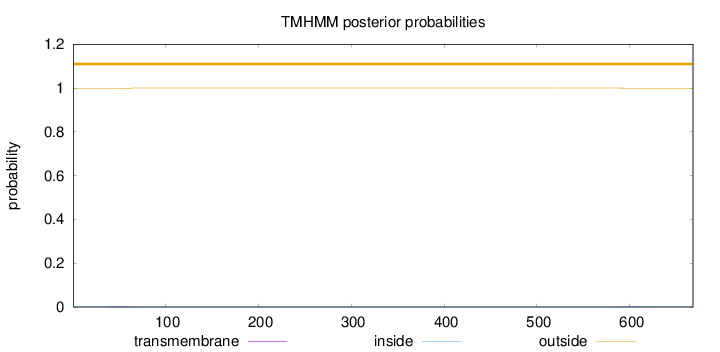
Topology
Length:
668
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05782
Exp number, first 60 AAs:
0.04116
Total prob of N-in:
0.00223
outside
1 - 668
Population Genetic Test Statistics
Pi
215.456041
Theta
156.002238
Tajima's D
1.278011
CLR
0.005204
CSRT
0.733963301834908
Interpretation
Uncertain
