Gene
KWMTBOMO03026
Annotation
PREDICTED:_piggyBac_transposable_element-derived_protein_4-like_isoform_X3_[Bombyx_mori]
Full name
PiggyBac transposable element-derived protein 4
Location in the cell
Nuclear Reliability : 1.768 PlasmaMembrane Reliability : 1.471
Sequence
CDS
ATGAACGTGTACCTTTTCCTTTCAGGCAGCGAAGAAGAGTCTGACGAAGAGGAAGAAGGGATTGCCGAGTTAGCGGTGCCCCGAATAGCAAGAATGTTTATGGACGAAGACGATGCGGAAGTCAACGAAAGATCCGCCAACGCTACTTTGGAGCGGTCCCCTCAATCGCCTCGACCAGCCGATTGTGCTTCAACGCCCGCCTCACCGACGCCTGTTTCGCCGACTGCCATGGAACCCAGCAACAAATTTCAATTTGAATGGAGAAAGTTTTCAACGCCTCAAATTGAACCTCATTTGAGGCGAGAACCGTTCTCGCAAATTAGTGGCCCTACAGTTTCTTTCACGAGACCATACGAAGCATTTATTGCTATATGGGATCGTGAAATTATGGAAATGATTGTCAGGGAAACTAACATTTACGCACAGCAGCTGGCAACGACCATGTTGGAGAATGGAACGATCTGTCCTAGTAGTCGAATTACTCGTTGGCATGATACAAATGTAAATGAACTTTACACGTATTTTGCCATCGTTCTGGCTATGGGAGTGGTGATCAAAACCCGGTTAACCGAATACTGGAATACCTCTAACGACATCTTCAGTACTCCTGGCTTTTCCACGGAGATGTCATATGACAGATTCCACCTCCTGTCAGCATGTCTGCATTTTAATAATAATAAAGATTGTAACTCTGCGTTGTTGACACGACCCCAAGCTAAACTTTTTAAAATTAAACCTATAATAGATCACCTTAATCGCAAGTTTTCTGAATTATATAGTTTGTCTCAAAATGTGGCTCTCGATGAGAGTCTGACCATGTGGAAGGGCTGGCTTGAAATAAACCAGTTCATCCCTAACAAAGCAGCCACAGTTGGAATCAAAACCTTTGAATTGTGTGAATCTCAGACTGGTTATTTGTGGCGTTTTGAAGTATATGCAGGCCATGACACCTCGGCATTTCAAGAAGATGATCCAGTCTCGGGGAAAGTACCAGCTCTAGTGCTCAGACTTTTAAATGGTCTTGAACATAAAGGGCACACAATATGGATGGACAATTACTACAACTCTCCATCACTGGCACGAGAACTGAAGATCCGGGGATTTGACTGCGCGGGGACGCTCCGGACAAACCGTCAATTCGTACCACCTGAATTGGCGAGTATTACAAAAAACGACATGATTGTCGGTCAATTGTTGGGATGCACCTCTGGAGATGTGGACATTGTGGTGTGGAGGGATAAAAACCTCGTGGCTTTTATTTCAACATACCATGCTCTCCTAGCTACCAAGTGTGGCGATACAGTTAAGCCGACAGTCGCGCAGGACTATAATCTTAATATGGGCGGAGTAGACCGTAAGGATCAGCAACTCGCAATGTACCCTTTAGAAAGAAAAAGGAATAAGGTGTGCGATGCTTATAAACATGATTAA
Protein
MNVYLFLSGSEEESDEEEEGIAELAVPRIARMFMDEDDAEVNERSANATLERSPQSPRPADCASTPASPTPVSPTAMEPSNKFQFEWRKFSTPQIEPHLRREPFSQISGPTVSFTRPYEAFIAIWDREIMEMIVRETNIYAQQLATTMLENGTICPSSRITRWHDTNVNELYTYFAIVLAMGVVIKTRLTEYWNTSNDIFSTPGFSTEMSYDRFHLLSACLHFNNNKDCNSALLTRPQAKLFKIKPIIDHLNRKFSELYSLSQNVALDESLTMWKGWLEINQFIPNKAATVGIKTFELCESQTGYLWRFEVYAGHDTSAFQEDDPVSGKVPALVLRLLNGLEHKGHTIWMDNYYNSPSLARELKIRGFDCAGTLRTNRQFVPPELASITKNDMIVGQLLGCTSGDVDIVVWRDKNLVAFISTYHALLATKCGDTVKPTVAQDYNLNMGGVDRKDQQLAMYPLERKRNKVCDAYKHD
Summary
Keywords
Complete proteome
Reference proteome
Feature
chain PiggyBac transposable element-derived protein 4
Uniprot
A0A194RRT0
A0A2H1V5F3
A0A2W1BDM1
A0A2A4IYX3
A0A2A4JR95
A0A2A4IY54
+ More
A0A2A4ITI3 A0A0L7L984 A0A2W1B9N8 A0A2J7PER0 A0A2A4K616 A0A2J7Q6H1 A0A2J7PD69 A0A2J7QZK8 A0A2J7PIK3 A0A2J7PRG6 A0A2J7QSN3 A0A2W1BNR3 A0A2J7PE61 A0A2J7Q8N9 A0A147BD88 F7FDY8 A0A2K6S7R1 A0A2K5PIR4 A0A2K5BXT0 H9F6U8 G7MWA2 G7PAN9 A0A2K5KZS5 Q96DM1 A0A2K6ALX5 G3QWT8 A0A2K6JV20 A0A2K6NHE6 A0A2K5UF88 A0A2K5HN84 A0A2K5XAW3 H2NMQ4 A0A0D9SEG0 A0A096NT85 A0A1D5R1S0 A0A1B6E873 K7DKZ9 A0A087TVE5 H9IWM5 A0A147BEZ3 A0A1B6LFF4 A0A087UJ51 A0A087TJF5 H9J421 A0A1B6H7X0 A0A0K0E1W0 J9LP86 A0A1B6LW93 A0A1B0FFI5 A0A1B6KKZ3 A0A2J7R6C2 A8J4K3 A0A3Q0JNJ9 A0A2J7Q4C7 A0A2J7PIT0 A0A2J7QF25 A0A1B6JYI5 A0A2J7PH34 A0A2J7R708 A0A1B6E8U4 A0A2J7QY13 A0A2J7R0C5 A0A2J7RD45 A8J4K9 H9J3V8 A0A2B4SRV7 E2AJF8 A0A1B6HI21 A0A2J7Q696 J9KLM1 A0A1A8PBD7 A0A3N0XE89 A0A023EYN9 A0A1A8D728 A0A1A8C9U5 A0A1A8PXY9 A8J4K6 A0A1B6KA50 A0A1B6FTB9 A0A1B6L7B9 T2M852 A0A1B6KBT3 A0A087TRV5 A0A1E1WHA1 A0A087U4Y0
A0A2A4ITI3 A0A0L7L984 A0A2W1B9N8 A0A2J7PER0 A0A2A4K616 A0A2J7Q6H1 A0A2J7PD69 A0A2J7QZK8 A0A2J7PIK3 A0A2J7PRG6 A0A2J7QSN3 A0A2W1BNR3 A0A2J7PE61 A0A2J7Q8N9 A0A147BD88 F7FDY8 A0A2K6S7R1 A0A2K5PIR4 A0A2K5BXT0 H9F6U8 G7MWA2 G7PAN9 A0A2K5KZS5 Q96DM1 A0A2K6ALX5 G3QWT8 A0A2K6JV20 A0A2K6NHE6 A0A2K5UF88 A0A2K5HN84 A0A2K5XAW3 H2NMQ4 A0A0D9SEG0 A0A096NT85 A0A1D5R1S0 A0A1B6E873 K7DKZ9 A0A087TVE5 H9IWM5 A0A147BEZ3 A0A1B6LFF4 A0A087UJ51 A0A087TJF5 H9J421 A0A1B6H7X0 A0A0K0E1W0 J9LP86 A0A1B6LW93 A0A1B0FFI5 A0A1B6KKZ3 A0A2J7R6C2 A8J4K3 A0A3Q0JNJ9 A0A2J7Q4C7 A0A2J7PIT0 A0A2J7QF25 A0A1B6JYI5 A0A2J7PH34 A0A2J7R708 A0A1B6E8U4 A0A2J7QY13 A0A2J7R0C5 A0A2J7RD45 A8J4K9 H9J3V8 A0A2B4SRV7 E2AJF8 A0A1B6HI21 A0A2J7Q696 J9KLM1 A0A1A8PBD7 A0A3N0XE89 A0A023EYN9 A0A1A8D728 A0A1A8C9U5 A0A1A8PXY9 A8J4K6 A0A1B6KA50 A0A1B6FTB9 A0A1B6L7B9 T2M852 A0A1B6KBT3 A0A087TRV5 A0A1E1WHA1 A0A087U4Y0
Pubmed
EMBL
KQ459989
KPJ18766.1
ODYU01000749
SOQ36006.1
KZ150113
PZC73302.1
+ More
NWSH01005160 PCG64390.1 NWSH01000726 PCG74565.1 NWSH01005605 PCG64100.1 NWSH01008683 PCG62453.1 JTDY01002247 KOB71806.1 KZ150197 PZC72319.1 NEVH01026090 PNF14815.1 NWSH01000091 PCG79681.1 NEVH01017475 NEVH01002684 NEVH01001351 PNF24176.1 PNF41738.1 PNF42927.1 NEVH01026393 PNF14286.1 NEVH01009069 PNF34004.1 NEVH01026401 NEVH01025126 NEVH01020854 NEVH01013255 NEVH01000629 PNF14193.1 PNF16176.1 PNF21085.1 PNF29260.1 PNF43158.1 NEVH01021985 PNF18924.1 NEVH01011876 PNF31583.1 PZC73303.1 NEVH01026109 PNF14626.1 NEVH01016950 PNF24933.1 GEGO01007089 JAR88315.1 JU326601 AFE70357.1 CM001259 EHH27175.1 CM001282 EHH62949.1 AK057200 AK094816 AK289491 AC079203 CABD030095328 AQIA01062119 ABGA01233763 NDHI03003689 PNJ08370.1 AQIB01116437 AHZZ02030739 JSUE03037923 GEDC01003168 JAS34130.1 AACZ04060380 GABC01006592 GABD01003812 GABE01001690 NBAG03000487 JAA04746.1 JAA29288.1 JAA43049.1 PNI20010.1 KK116918 KFM69084.1 BABH01021160 BABH01021161 BABH01021162 BABH01021163 GEGO01006047 JAR89357.1 GEBQ01017560 JAT22417.1 KK120039 KFM77390.1 KK115494 KFM65244.1 BABH01041064 GECU01037003 GECU01013756 JAS70703.1 JAS93950.1 ABLF02006169 ABLF02055685 GEBQ01012026 JAT27951.1 CCAG010009903 GEBQ01027849 JAT12128.1 NEVH01006795 PNF36382.1 AB300556 BAF82019.1 NEVH01018384 PNF23441.1 NEVH01024959 PNF16251.1 NEVH01015302 PNF27192.1 GECU01003439 JAT04268.1 NEVH01025150 PNF15655.1 NEVH01006740 PNF36596.1 GEDC01002971 JAS34327.1 NEVH01009368 PNF33480.1 NEVH01008275 PNF34265.1 NEVH01005303 PNF38748.1 AB300558 BAF82021.1 BABH01028864 LSMT01000021 PFX32631.1 GL439981 EFN66430.1 GECU01033435 GECU01003618 JAS74271.1 JAT04089.1 NEVH01017534 PNF24104.1 ABLF02028028 HAEI01002033 SBR78690.1 RJVU01079141 ROI15640.1 GBBI01004439 JAC14273.1 HAEA01000274 SBQ28754.1 HADZ01012453 SBP76394.1 HAEH01008939 SBR85849.1 AB300557 BAF82020.1 GEBQ01031648 JAT08329.1 GECZ01016293 JAS53476.1 GEBQ01020417 GEBQ01019545 JAT19560.1 JAT20432.1 HAAD01001920 CDG68152.1 GEBQ01031067 JAT08910.1 KK116466 KFM67844.1 GDQN01004768 JAT86286.1 KK118194 KFM72419.1
NWSH01005160 PCG64390.1 NWSH01000726 PCG74565.1 NWSH01005605 PCG64100.1 NWSH01008683 PCG62453.1 JTDY01002247 KOB71806.1 KZ150197 PZC72319.1 NEVH01026090 PNF14815.1 NWSH01000091 PCG79681.1 NEVH01017475 NEVH01002684 NEVH01001351 PNF24176.1 PNF41738.1 PNF42927.1 NEVH01026393 PNF14286.1 NEVH01009069 PNF34004.1 NEVH01026401 NEVH01025126 NEVH01020854 NEVH01013255 NEVH01000629 PNF14193.1 PNF16176.1 PNF21085.1 PNF29260.1 PNF43158.1 NEVH01021985 PNF18924.1 NEVH01011876 PNF31583.1 PZC73303.1 NEVH01026109 PNF14626.1 NEVH01016950 PNF24933.1 GEGO01007089 JAR88315.1 JU326601 AFE70357.1 CM001259 EHH27175.1 CM001282 EHH62949.1 AK057200 AK094816 AK289491 AC079203 CABD030095328 AQIA01062119 ABGA01233763 NDHI03003689 PNJ08370.1 AQIB01116437 AHZZ02030739 JSUE03037923 GEDC01003168 JAS34130.1 AACZ04060380 GABC01006592 GABD01003812 GABE01001690 NBAG03000487 JAA04746.1 JAA29288.1 JAA43049.1 PNI20010.1 KK116918 KFM69084.1 BABH01021160 BABH01021161 BABH01021162 BABH01021163 GEGO01006047 JAR89357.1 GEBQ01017560 JAT22417.1 KK120039 KFM77390.1 KK115494 KFM65244.1 BABH01041064 GECU01037003 GECU01013756 JAS70703.1 JAS93950.1 ABLF02006169 ABLF02055685 GEBQ01012026 JAT27951.1 CCAG010009903 GEBQ01027849 JAT12128.1 NEVH01006795 PNF36382.1 AB300556 BAF82019.1 NEVH01018384 PNF23441.1 NEVH01024959 PNF16251.1 NEVH01015302 PNF27192.1 GECU01003439 JAT04268.1 NEVH01025150 PNF15655.1 NEVH01006740 PNF36596.1 GEDC01002971 JAS34327.1 NEVH01009368 PNF33480.1 NEVH01008275 PNF34265.1 NEVH01005303 PNF38748.1 AB300558 BAF82021.1 BABH01028864 LSMT01000021 PFX32631.1 GL439981 EFN66430.1 GECU01033435 GECU01003618 JAS74271.1 JAT04089.1 NEVH01017534 PNF24104.1 ABLF02028028 HAEI01002033 SBR78690.1 RJVU01079141 ROI15640.1 GBBI01004439 JAC14273.1 HAEA01000274 SBQ28754.1 HADZ01012453 SBP76394.1 HAEH01008939 SBR85849.1 AB300557 BAF82020.1 GEBQ01031648 JAT08329.1 GECZ01016293 JAS53476.1 GEBQ01020417 GEBQ01019545 JAT19560.1 JAT20432.1 HAAD01001920 CDG68152.1 GEBQ01031067 JAT08910.1 KK116466 KFM67844.1 GDQN01004768 JAT86286.1 KK118194 KFM72419.1
Proteomes
UP000053240
UP000218220
UP000037510
UP000235965
UP000008225
UP000233220
+ More
UP000233040 UP000233020 UP000009130 UP000233060 UP000005640 UP000233120 UP000001519 UP000233180 UP000233200 UP000233100 UP000233080 UP000233140 UP000001595 UP000029965 UP000028761 UP000006718 UP000002277 UP000054359 UP000005204 UP000035681 UP000007819 UP000092444 UP000079169 UP000225706 UP000000311
UP000233040 UP000233020 UP000009130 UP000233060 UP000005640 UP000233120 UP000001519 UP000233180 UP000233200 UP000233100 UP000233080 UP000233140 UP000001595 UP000029965 UP000028761 UP000006718 UP000002277 UP000054359 UP000005204 UP000035681 UP000007819 UP000092444 UP000079169 UP000225706 UP000000311
Pfam
Interpro
ProteinModelPortal
A0A194RRT0
A0A2H1V5F3
A0A2W1BDM1
A0A2A4IYX3
A0A2A4JR95
A0A2A4IY54
+ More
A0A2A4ITI3 A0A0L7L984 A0A2W1B9N8 A0A2J7PER0 A0A2A4K616 A0A2J7Q6H1 A0A2J7PD69 A0A2J7QZK8 A0A2J7PIK3 A0A2J7PRG6 A0A2J7QSN3 A0A2W1BNR3 A0A2J7PE61 A0A2J7Q8N9 A0A147BD88 F7FDY8 A0A2K6S7R1 A0A2K5PIR4 A0A2K5BXT0 H9F6U8 G7MWA2 G7PAN9 A0A2K5KZS5 Q96DM1 A0A2K6ALX5 G3QWT8 A0A2K6JV20 A0A2K6NHE6 A0A2K5UF88 A0A2K5HN84 A0A2K5XAW3 H2NMQ4 A0A0D9SEG0 A0A096NT85 A0A1D5R1S0 A0A1B6E873 K7DKZ9 A0A087TVE5 H9IWM5 A0A147BEZ3 A0A1B6LFF4 A0A087UJ51 A0A087TJF5 H9J421 A0A1B6H7X0 A0A0K0E1W0 J9LP86 A0A1B6LW93 A0A1B0FFI5 A0A1B6KKZ3 A0A2J7R6C2 A8J4K3 A0A3Q0JNJ9 A0A2J7Q4C7 A0A2J7PIT0 A0A2J7QF25 A0A1B6JYI5 A0A2J7PH34 A0A2J7R708 A0A1B6E8U4 A0A2J7QY13 A0A2J7R0C5 A0A2J7RD45 A8J4K9 H9J3V8 A0A2B4SRV7 E2AJF8 A0A1B6HI21 A0A2J7Q696 J9KLM1 A0A1A8PBD7 A0A3N0XE89 A0A023EYN9 A0A1A8D728 A0A1A8C9U5 A0A1A8PXY9 A8J4K6 A0A1B6KA50 A0A1B6FTB9 A0A1B6L7B9 T2M852 A0A1B6KBT3 A0A087TRV5 A0A1E1WHA1 A0A087U4Y0
A0A2A4ITI3 A0A0L7L984 A0A2W1B9N8 A0A2J7PER0 A0A2A4K616 A0A2J7Q6H1 A0A2J7PD69 A0A2J7QZK8 A0A2J7PIK3 A0A2J7PRG6 A0A2J7QSN3 A0A2W1BNR3 A0A2J7PE61 A0A2J7Q8N9 A0A147BD88 F7FDY8 A0A2K6S7R1 A0A2K5PIR4 A0A2K5BXT0 H9F6U8 G7MWA2 G7PAN9 A0A2K5KZS5 Q96DM1 A0A2K6ALX5 G3QWT8 A0A2K6JV20 A0A2K6NHE6 A0A2K5UF88 A0A2K5HN84 A0A2K5XAW3 H2NMQ4 A0A0D9SEG0 A0A096NT85 A0A1D5R1S0 A0A1B6E873 K7DKZ9 A0A087TVE5 H9IWM5 A0A147BEZ3 A0A1B6LFF4 A0A087UJ51 A0A087TJF5 H9J421 A0A1B6H7X0 A0A0K0E1W0 J9LP86 A0A1B6LW93 A0A1B0FFI5 A0A1B6KKZ3 A0A2J7R6C2 A8J4K3 A0A3Q0JNJ9 A0A2J7Q4C7 A0A2J7PIT0 A0A2J7QF25 A0A1B6JYI5 A0A2J7PH34 A0A2J7R708 A0A1B6E8U4 A0A2J7QY13 A0A2J7R0C5 A0A2J7RD45 A8J4K9 H9J3V8 A0A2B4SRV7 E2AJF8 A0A1B6HI21 A0A2J7Q696 J9KLM1 A0A1A8PBD7 A0A3N0XE89 A0A023EYN9 A0A1A8D728 A0A1A8C9U5 A0A1A8PXY9 A8J4K6 A0A1B6KA50 A0A1B6FTB9 A0A1B6L7B9 T2M852 A0A1B6KBT3 A0A087TRV5 A0A1E1WHA1 A0A087U4Y0
Ontologies
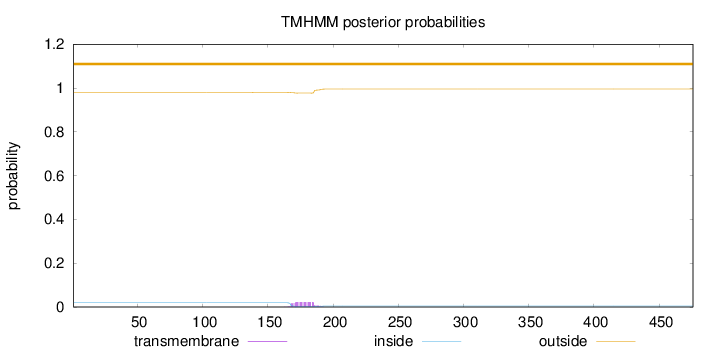
Topology
Subcellular location
Nucleus
Length:
476
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43064
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01947
outside
1 - 476
Population Genetic Test Statistics
Pi
417.256773
Theta
235.474135
Tajima's D
3.202493
CLR
0.514347
CSRT
0.985350732463377
Interpretation
Possibly Positive selection
