Gene
KWMTBOMO03025
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.545 Nuclear Reliability : 2.012
Sequence
CDS
ATGACTTACGGTTCCGAGACGTGGTGCTTCACAAGGGATCTTTTCAGGAAGCTTAGTGTCGCGCAGCGAGCTATGGAGAGGGCTATGCTTGGTATTTCTCTACGAGATCGAATCAGAAATGAGGAGATCCGTAGAAGAACCAAAGTTGCTGACATAGCCCGTAGGATTAGCAAGCTGAAGTGGCAGTGGGCTGGCCACATAGCGCGAAGATCGGACGGCCGATGGGGCAGAAAGATCCTCGAGTGGAGACCACGTACTGGCAAACGCAGTGTGGGACGGCCACCTACCAGATGGACCGACGACCTGGTGAAAATTGCTGGGTCACGTTGGATGCAGGTGGCAACGGACCGGCCGCACTGGACATCAGTTGGAGAGGCCTATGTTCAGCAGTGGACAGAAGACAGGCTGAAATGA
Protein
MTYGSETWCFTRDLFRKLSVAQRAMERAMLGISLRDRIRNEEIRRRTKVADIARRISKLKWQWAGHIARRSDGRWGRKILEWRPRTGKRSVGRPPTRWTDDLVKIAGSRWMQVATDRPHWTSVGEAYVQQWTEDRLK
Summary
Uniprot
D7F159
D7F158
A4KWF4
A4KWF2
A4KWG0
D7F164
+ More
D7F157 A0A2W1BMB6 D7F160 D7F166 D7F165 A0A2W1BGB6 A0A2H1WG43 A0A2W1BSC0 A0A2H1WM45 A0A2H1WXR7 A0A2H1VRK2 A0A2W1BLC9 A0A023EXV2 A0A2W1BCS2 A0A2A4J310 D7F179 D7F178 A0A2W1B691 A0A131YK20 E2AL50 A0A0B2US56 D7F176 A0A0B2VLG9 A0A2G8K8G7 A0A0J7KXZ5 A0A0B2UXX4 A0A2G8JFM9 A0A2W1B5F8 A0A2H1V663 A0A2W1BDX2 A0A131Z9K5 A0A2A4J025 A0A2G8JIM1 D7F177 W4YEN5 A0A2W1BSQ8 A0A0B2VA05 A0A023EWU8 W4XDK0 A0A0P4VWD0 W4YJQ5 A0A0B2VKB7 W4XCB7 W4YXJ1 W4YTY1 A0A2G8K4F2 A0A016VZE9 A0A016VZ67 A0A1E1XN17 A0A069DQ34 A0A2H1VY30 A0A224YVW5 W4YRL2 A0A183UMK8 A0A069DX96 A0A016W5P9 A0A2H1W4U6 E3NGJ2 A0A016SG46 A0A016W4R1 A0A016TK11 E3NC15 A0A2H1WE04 W4XMW4 E3MKV4 E3NQY5 E3LKV7 E3NNL1 E3N0F4 E3MII9 E3NMS2 E3NKD0 E3MH09 W4YU42 E3NB36 E3NSE7
D7F157 A0A2W1BMB6 D7F160 D7F166 D7F165 A0A2W1BGB6 A0A2H1WG43 A0A2W1BSC0 A0A2H1WM45 A0A2H1WXR7 A0A2H1VRK2 A0A2W1BLC9 A0A023EXV2 A0A2W1BCS2 A0A2A4J310 D7F179 D7F178 A0A2W1B691 A0A131YK20 E2AL50 A0A0B2US56 D7F176 A0A0B2VLG9 A0A2G8K8G7 A0A0J7KXZ5 A0A0B2UXX4 A0A2G8JFM9 A0A2W1B5F8 A0A2H1V663 A0A2W1BDX2 A0A131Z9K5 A0A2A4J025 A0A2G8JIM1 D7F177 W4YEN5 A0A2W1BSQ8 A0A0B2VA05 A0A023EWU8 W4XDK0 A0A0P4VWD0 W4YJQ5 A0A0B2VKB7 W4XCB7 W4YXJ1 W4YTY1 A0A2G8K4F2 A0A016VZE9 A0A016VZ67 A0A1E1XN17 A0A069DQ34 A0A2H1VY30 A0A224YVW5 W4YRL2 A0A183UMK8 A0A069DX96 A0A016W5P9 A0A2H1W4U6 E3NGJ2 A0A016SG46 A0A016W4R1 A0A016TK11 E3NC15 A0A2H1WE04 W4XMW4 E3MKV4 E3NQY5 E3LKV7 E3NNL1 E3N0F4 E3MII9 E3NMS2 E3NKD0 E3MH09 W4YU42 E3NB36 E3NSE7
Pubmed
EMBL
FJ265544
ADI61812.1
FJ265543
ADI61811.1
EF113399
EF113400
+ More
ABO45233.1 EF113398 EF113401 ABO45231.1 EF113402 ABO45239.1 FJ265549 ADI61817.1 FJ265542 ADI61810.1 KZ150072 PZC74016.1 FJ265545 ADI61813.1 FJ265551 ADI61819.1 FJ265550 ADI61818.1 KZ150065 PZC74169.1 ODYU01008431 SOQ52031.1 KZ149953 PZC76544.1 ODYU01009605 SOQ54153.1 ODYU01011862 SOQ57848.1 ODYU01003998 SOQ43428.1 KZ149985 PZC75698.1 GBBI01004876 JAC13836.1 KZ150485 PZC70710.1 NWSH01003335 PCG66527.1 FJ265564 ADI61832.1 FJ265563 ADI61831.1 KZ150286 PZC71609.1 GEDV01010296 JAP78261.1 GL440455 EFN65840.1 JPKZ01004123 KHN71972.1 FJ265561 ADI61829.1 JPKZ01001394 KHN82224.1 MRZV01000786 PIK44294.1 LBMM01002200 KMQ95153.1 JPKZ01002932 KHN74308.1 MRZV01002136 PIK34564.1 KZ150317 PZC71439.1 ODYU01000693 SOQ35892.1 KZ150108 PZC73382.1 GEDV01000518 JAP88039.1 NWSH01004509 PCG65028.1 MRZV01001865 PIK35593.1 FJ265562 ADI61830.1 AAGJ04035986 KZ149912 PZC78089.1 JPKZ01002136 KHN78309.1 GBBI01004877 JAC13835.1 AAGJ04011852 GDKW01001596 JAI54999.1 AAGJ04005502 JPKZ01001456 KHN81799.1 AAGJ04065374 AAGJ04144474 AAGJ04012759 MRZV01000899 PIK42833.1 JARK01001338 EYC32701.1 EYC32700.1 GFAA01002978 JAU00457.1 GBGD01002934 JAC85955.1 ODYU01004982 SOQ45402.1 GFPF01007585 MAA18731.1 AAGJ04112759 UYWY01020264 VDM41049.1 GBGD01000453 JAC88436.1 JARK01000732 EYC35149.1 ODYU01006342 SOQ48110.1 DS268655 EFO97114.1 JARK01001565 EYB89683.1 JARK01001337 EYC34625.1 JARK01001676 JARK01001446 JARK01001433 EYB83199.1 EYC01126.1 EYC02928.1 DS268591 EFO92565.1 ODYU01008030 SOQ51293.1 AAGJ04013248 DS268453 EFP04265.1 DS269613 EFO86468.1 DS268410 EFO99851.1 DS269226 EFP10832.1 DS268505 EFP13372.1 DS268448 EFP02560.1 DS269097 EFP08676.1 DS268794 EFP02105.1 DS268444 EFP01729.1 AAGJ04112537 DS268581 EFO91764.1 DS269943 EFO90109.1
ABO45233.1 EF113398 EF113401 ABO45231.1 EF113402 ABO45239.1 FJ265549 ADI61817.1 FJ265542 ADI61810.1 KZ150072 PZC74016.1 FJ265545 ADI61813.1 FJ265551 ADI61819.1 FJ265550 ADI61818.1 KZ150065 PZC74169.1 ODYU01008431 SOQ52031.1 KZ149953 PZC76544.1 ODYU01009605 SOQ54153.1 ODYU01011862 SOQ57848.1 ODYU01003998 SOQ43428.1 KZ149985 PZC75698.1 GBBI01004876 JAC13836.1 KZ150485 PZC70710.1 NWSH01003335 PCG66527.1 FJ265564 ADI61832.1 FJ265563 ADI61831.1 KZ150286 PZC71609.1 GEDV01010296 JAP78261.1 GL440455 EFN65840.1 JPKZ01004123 KHN71972.1 FJ265561 ADI61829.1 JPKZ01001394 KHN82224.1 MRZV01000786 PIK44294.1 LBMM01002200 KMQ95153.1 JPKZ01002932 KHN74308.1 MRZV01002136 PIK34564.1 KZ150317 PZC71439.1 ODYU01000693 SOQ35892.1 KZ150108 PZC73382.1 GEDV01000518 JAP88039.1 NWSH01004509 PCG65028.1 MRZV01001865 PIK35593.1 FJ265562 ADI61830.1 AAGJ04035986 KZ149912 PZC78089.1 JPKZ01002136 KHN78309.1 GBBI01004877 JAC13835.1 AAGJ04011852 GDKW01001596 JAI54999.1 AAGJ04005502 JPKZ01001456 KHN81799.1 AAGJ04065374 AAGJ04144474 AAGJ04012759 MRZV01000899 PIK42833.1 JARK01001338 EYC32701.1 EYC32700.1 GFAA01002978 JAU00457.1 GBGD01002934 JAC85955.1 ODYU01004982 SOQ45402.1 GFPF01007585 MAA18731.1 AAGJ04112759 UYWY01020264 VDM41049.1 GBGD01000453 JAC88436.1 JARK01000732 EYC35149.1 ODYU01006342 SOQ48110.1 DS268655 EFO97114.1 JARK01001565 EYB89683.1 JARK01001337 EYC34625.1 JARK01001676 JARK01001446 JARK01001433 EYB83199.1 EYC01126.1 EYC02928.1 DS268591 EFO92565.1 ODYU01008030 SOQ51293.1 AAGJ04013248 DS268453 EFP04265.1 DS269613 EFO86468.1 DS268410 EFO99851.1 DS269226 EFP10832.1 DS268505 EFP13372.1 DS268448 EFP02560.1 DS269097 EFP08676.1 DS268794 EFP02105.1 DS268444 EFP01729.1 AAGJ04112537 DS268581 EFO91764.1 DS269943 EFO90109.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D7F159
D7F158
A4KWF4
A4KWF2
A4KWG0
D7F164
+ More
D7F157 A0A2W1BMB6 D7F160 D7F166 D7F165 A0A2W1BGB6 A0A2H1WG43 A0A2W1BSC0 A0A2H1WM45 A0A2H1WXR7 A0A2H1VRK2 A0A2W1BLC9 A0A023EXV2 A0A2W1BCS2 A0A2A4J310 D7F179 D7F178 A0A2W1B691 A0A131YK20 E2AL50 A0A0B2US56 D7F176 A0A0B2VLG9 A0A2G8K8G7 A0A0J7KXZ5 A0A0B2UXX4 A0A2G8JFM9 A0A2W1B5F8 A0A2H1V663 A0A2W1BDX2 A0A131Z9K5 A0A2A4J025 A0A2G8JIM1 D7F177 W4YEN5 A0A2W1BSQ8 A0A0B2VA05 A0A023EWU8 W4XDK0 A0A0P4VWD0 W4YJQ5 A0A0B2VKB7 W4XCB7 W4YXJ1 W4YTY1 A0A2G8K4F2 A0A016VZE9 A0A016VZ67 A0A1E1XN17 A0A069DQ34 A0A2H1VY30 A0A224YVW5 W4YRL2 A0A183UMK8 A0A069DX96 A0A016W5P9 A0A2H1W4U6 E3NGJ2 A0A016SG46 A0A016W4R1 A0A016TK11 E3NC15 A0A2H1WE04 W4XMW4 E3MKV4 E3NQY5 E3LKV7 E3NNL1 E3N0F4 E3MII9 E3NMS2 E3NKD0 E3MH09 W4YU42 E3NB36 E3NSE7
D7F157 A0A2W1BMB6 D7F160 D7F166 D7F165 A0A2W1BGB6 A0A2H1WG43 A0A2W1BSC0 A0A2H1WM45 A0A2H1WXR7 A0A2H1VRK2 A0A2W1BLC9 A0A023EXV2 A0A2W1BCS2 A0A2A4J310 D7F179 D7F178 A0A2W1B691 A0A131YK20 E2AL50 A0A0B2US56 D7F176 A0A0B2VLG9 A0A2G8K8G7 A0A0J7KXZ5 A0A0B2UXX4 A0A2G8JFM9 A0A2W1B5F8 A0A2H1V663 A0A2W1BDX2 A0A131Z9K5 A0A2A4J025 A0A2G8JIM1 D7F177 W4YEN5 A0A2W1BSQ8 A0A0B2VA05 A0A023EWU8 W4XDK0 A0A0P4VWD0 W4YJQ5 A0A0B2VKB7 W4XCB7 W4YXJ1 W4YTY1 A0A2G8K4F2 A0A016VZE9 A0A016VZ67 A0A1E1XN17 A0A069DQ34 A0A2H1VY30 A0A224YVW5 W4YRL2 A0A183UMK8 A0A069DX96 A0A016W5P9 A0A2H1W4U6 E3NGJ2 A0A016SG46 A0A016W4R1 A0A016TK11 E3NC15 A0A2H1WE04 W4XMW4 E3MKV4 E3NQY5 E3LKV7 E3NNL1 E3N0F4 E3MII9 E3NMS2 E3NKD0 E3MH09 W4YU42 E3NB36 E3NSE7
Ontologies
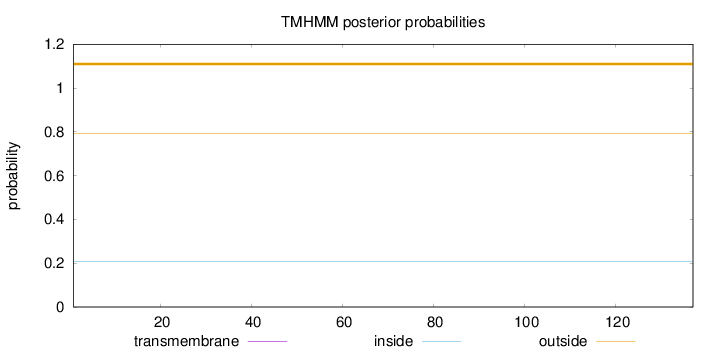
Topology
Length:
137
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00034
Exp number, first 60 AAs:
0.00034
Total prob of N-in:
0.20821
outside
1 - 137
Population Genetic Test Statistics
Pi
329.128172
Theta
183.911586
Tajima's D
2.484541
CLR
0
CSRT
0.941402929853507
Interpretation
Uncertain
