Gene
KWMTBOMO03019
Pre Gene Modal
BGIBMGA003601
Annotation
PREDICTED:_beta-arrestin-1_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.546
Sequence
CDS
ATGTCTGCGATCACAGTGTATTTGGGAAAGAGGGATTTCGTCGATCACATTTCACACGTAGACCCGATCGATGGGGTCGTACTAATAGATCCGGAATACGTGAAAGATAGAAAGGTTTTTGGTCATGTCTTGGCGGCGTTCAGGTACGGACGGGAGGACTTGGACGTGCTCGGACTTACGTTCAGGAAGGATCTGTATTTAGCCGCCGAACAGATATTCCCGCCGACGAGCACGGTAAAGCGACCGCTGACCCGACTACAAGAACGCTTAGTCCGCAAACTGGGGCCATCGGCTCACCCGTTCTACTTCGAGCTGCCTCCGCACTGTCCGGCCTCGGTCACCCTGCAGCCGGCTCCGGGAGACACCGGCAAGCCTTGCGGAGTGGACTATGAGCTTAAAGCGTTCGTCGCCGATTCCCAAGATGACAAACCCCACAAAAGGAATTCGGTTCGGTTAGCAATCAGGAAGATAATGTACGCGCCGAGCAAGCAAGGCGAACAGCCTTCAGTCGAAGTCTCGAAAGAGTTCATGATGAGCCCTAACAAACTGTATCTTGAGGCATCTTTAGATAAGGAGCTTTACTACCACGGCGAGAACATAGCGGTGAACGTGCACATAGCGAACAACTCGAATCGATCTGTGAAGCGCATCAAGGTGTCGGTGCGGCAGTTCGCCGACATTTGCCTCTTCTCCACCGCCCAGTACAAGTGCACCGTCGCCGAGGCGGAGAGCGAGGAGGGTTGTCCCGTGGGACCTGGGTTCACGCTCAGCAAGGTGTTCACGCTGACGCCGCTGCTCGCCAACAATAAGGACAAGTGGGGCTTGGCGCTTGACGGACAACTCAAACACGAGGACACGAACCTGGCCTCCAGCACCCTTATAGCTGATCCGGCGCAACGAGAAAATTTGGGTATCATAGTTCAATACAAAGTTAAAGTGAAGCTGTGCTTGGGACCGCTCGGAGGTGAGCTGAGCGCCGAGCTGCCGTTCATCCTGATGCATCCGAAGCCCGAGGAGGAGCCGCGCGCGCCCGACCAGCCCCACGCGACCCCCGACCACGACCTCATCCAACTCGACCCGCCTCCAGATGAAAACGGCCAAGAACAAGATGACGATATAATATTCGAAGACTTCGCTCGACTCCGACTGAAGGGCGCCGACGCAGACGCCTGA
Protein
MSAITVYLGKRDFVDHISHVDPIDGVVLIDPEYVKDRKVFGHVLAAFRYGREDLDVLGLTFRKDLYLAAEQIFPPTSTVKRPLTRLQERLVRKLGPSAHPFYFELPPHCPASVTLQPAPGDTGKPCGVDYELKAFVADSQDDKPHKRNSVRLAIRKIMYAPSKQGEQPSVEVSKEFMMSPNKLYLEASLDKELYYHGENIAVNVHIANNSNRSVKRIKVSVRQFADICLFSTAQYKCTVAEAESEEGCPVGPGFTLSKVFTLTPLLANNKDKWGLALDGQLKHEDTNLASSTLIADPAQRENLGIIVQYKVKVKLCLGPLGGELSAELPFILMHPKPEEEPRAPDQPHATPDHDLIQLDPPPDENGQEQDDDIIFEDFARLRLKGADADA
Summary
Uniprot
M1Q038
A0A2H1VRW2
A0A194PQ77
A0A0N1ID58
A0A0C5B7N5
A0A2A4K8U9
+ More
H9J264 A0A212EWT8 V5I9D5 A0A1B6F7C9 A0A1Y1N6J5 A0A2J7PCY8 D6W6M4 A0A023F6Q2 A0A1B6EHG1 A0A1B6IW07 A0A1B6LNF2 A0A0A9XV02 A0A0P4VWJ9 A0A2J7PCY3 A0A1B6G1Z7 A0A0V0GBD4 A0A224XCZ1 A0A067RMV7 A0A125S9L8 A0A2H4WAL5 A0A1W4XPX9 A0A0A9Y0J1 A0A1L8DMN7 V5GVA6 T1IZ20 A0A0N7ZDG5 T1E2L5 A0A1L8DMN8 A0A2M4CTC7 A0A2M3Z5H1 A0A2M3Z5I6 A0A2R5LMJ0 A0A154PP49 A0A226ER22 A0A3L8DKJ2 A0A2M4A446 E2C2N9 B0XDH4 A0A026WGJ5 A0A182Y727 A0A1Q3F422 A0A293LZZ7 A0A1S4H7B9 Q66GU5 Q7Q2V9 A0A1J1J6H2 A0A195AZS5 U5EP45 A0A023G4Y8 G3MGZ3 A0A195CVL7 A0A0C9R0P7 A0A158NMH5 A0A151JSD1 F4WI29 A0A023FMD3 A0A151WUS5 A0A151JVY0 E2ASV7 V9IAX1 A0A131Z1I8 A0A224YT92 A0A2A3EQU3 A0A087ZUT4 A0A0M9ACM6 A0A182FPL5 Q16NE5 T1KCR5 T1KRS3 A0A2M3Z5U6 A0A084WAI6 A0A182Q7L7 A0A182PGH6 A0A182KK83 A0A2H8TQN5 A0A2S2NGI2 X1X3E0 A0A1Z5L0B8 A0A0P6FT56 A0A182RVF8 A0A0P6I3D5 A0A164RB02 B4PP42 A0A1J1J4N9 A0A1B0C8T4 A0A2C9GQ18 A0A0K2V853 E9HH60 B4K866 A0A0L0CJ44 A0A1A9Z4B4 T1PM66 A0A1B0FL93 A0A1B0AV75
H9J264 A0A212EWT8 V5I9D5 A0A1B6F7C9 A0A1Y1N6J5 A0A2J7PCY8 D6W6M4 A0A023F6Q2 A0A1B6EHG1 A0A1B6IW07 A0A1B6LNF2 A0A0A9XV02 A0A0P4VWJ9 A0A2J7PCY3 A0A1B6G1Z7 A0A0V0GBD4 A0A224XCZ1 A0A067RMV7 A0A125S9L8 A0A2H4WAL5 A0A1W4XPX9 A0A0A9Y0J1 A0A1L8DMN7 V5GVA6 T1IZ20 A0A0N7ZDG5 T1E2L5 A0A1L8DMN8 A0A2M4CTC7 A0A2M3Z5H1 A0A2M3Z5I6 A0A2R5LMJ0 A0A154PP49 A0A226ER22 A0A3L8DKJ2 A0A2M4A446 E2C2N9 B0XDH4 A0A026WGJ5 A0A182Y727 A0A1Q3F422 A0A293LZZ7 A0A1S4H7B9 Q66GU5 Q7Q2V9 A0A1J1J6H2 A0A195AZS5 U5EP45 A0A023G4Y8 G3MGZ3 A0A195CVL7 A0A0C9R0P7 A0A158NMH5 A0A151JSD1 F4WI29 A0A023FMD3 A0A151WUS5 A0A151JVY0 E2ASV7 V9IAX1 A0A131Z1I8 A0A224YT92 A0A2A3EQU3 A0A087ZUT4 A0A0M9ACM6 A0A182FPL5 Q16NE5 T1KCR5 T1KRS3 A0A2M3Z5U6 A0A084WAI6 A0A182Q7L7 A0A182PGH6 A0A182KK83 A0A2H8TQN5 A0A2S2NGI2 X1X3E0 A0A1Z5L0B8 A0A0P6FT56 A0A182RVF8 A0A0P6I3D5 A0A164RB02 B4PP42 A0A1J1J4N9 A0A1B0C8T4 A0A2C9GQ18 A0A0K2V853 E9HH60 B4K866 A0A0L0CJ44 A0A1A9Z4B4 T1PM66 A0A1B0FL93 A0A1B0AV75
Pubmed
26354079
25660147
19121390
22118469
28004739
18362917
+ More
19820115 25474469 25401762 26823975 27129103 24845553 26846853 29182505 25765539 24330624 30249741 20798317 24508170 25244985 12364791 14986925 22216098 21347285 21719571 26830274 28797301 17510324 24438588 20966253 28528879 17994087 17550304 21292972 26108605 25315136
19820115 25474469 25401762 26823975 27129103 24845553 26846853 29182505 25765539 24330624 30249741 20798317 24508170 25244985 12364791 14986925 22216098 21347285 21719571 26830274 28797301 17510324 24438588 20966253 28528879 17994087 17550304 21292972 26108605 25315136
EMBL
KC167880
AGF86399.1
ODYU01004061
SOQ43547.1
KQ459597
KPI95118.1
+ More
KQ461187 KPJ07358.1 KP027422 AJL99442.1 NWSH01000041 PCG80338.1 BABH01007386 BABH01007387 AGBW02011913 OWR45931.1 GALX01003125 JAB65341.1 GECZ01023669 JAS46100.1 GEZM01013441 GEZM01013440 GEZM01013439 GEZM01013438 GEZM01013437 GEZM01013436 GEZM01013435 GEZM01013434 JAV92550.1 NEVH01026401 PNF14195.1 KQ971307 EFA11544.2 GBBI01001802 JAC16910.1 GECZ01032403 GECZ01006964 JAS37366.1 JAS62805.1 GECU01019070 GECU01016580 GECU01011865 JAS88636.1 JAS91126.1 JAS95841.1 GEBQ01014715 JAT25262.1 GBHO01019003 GBHO01019002 GDHC01014969 GDHC01014142 JAG24601.1 JAG24602.1 JAQ03660.1 JAQ04487.1 GDKW01002630 JAI53965.1 PNF14194.1 GECZ01021874 GECZ01013303 JAS47895.1 JAS56466.1 GECL01000821 JAP05303.1 GFTR01006110 JAW10316.1 KK852415 KDR24393.1 KU160500 AME17864.1 MG020525 AUC64081.1 GBHO01019001 GBHO01013057 JAG24603.1 JAG30547.1 GFDF01006447 JAV07637.1 GANP01010138 JAB74330.1 JH431704 GDRN01039794 GDRN01039793 JAI67184.1 GALA01001155 JAA93697.1 GFDF01006372 JAV07712.1 GGFL01004351 MBW68529.1 GGFM01003020 MBW23771.1 GGFM01003025 MBW23776.1 GGLE01006598 MBY10724.1 KQ435007 KZC13665.1 LNIX01000002 OXA59648.1 QOIP01000007 RLU20964.1 GGFK01002208 MBW35529.1 GL452201 EFN77795.1 DS232757 EDS45452.1 KK107238 EZA54816.1 GFDL01012807 JAV22238.1 GFWV01008619 MAA33348.1 AAAB01008966 BK000997 DAA00889.1 EAA13078.1 CVRI01000070 CRL07380.1 KQ976697 KYM77542.1 GANO01000322 JAB59549.1 GBBM01006544 JAC28874.1 JO841144 AEO32761.1 KQ977231 KYN04721.1 GBYB01009714 JAG79481.1 ADTU01020500 ADTU01020501 ADTU01020502 ADTU01020503 KQ978577 KYN30051.1 GL888172 EGI66122.1 GBBK01002492 JAC21990.1 KQ982727 KYQ51603.1 KQ981676 KYN38147.1 GL442363 EFN63489.1 JR037128 AEY57596.1 GEDV01003769 JAP84788.1 GFPF01006337 MAA17483.1 KZ288193 PBC34135.1 KQ435701 KOX80419.1 CH477828 EAT35854.1 CAEY01001996 CAEY01000418 GGFM01003077 MBW23828.1 ATLV01022194 KE525329 KFB47230.1 AXCN02001120 GFXV01004698 MBW16503.1 GGMR01003646 MBY16265.1 ABLF02027074 GFJQ02006144 JAW00826.1 GDIQ01185918 GDIQ01133752 GDIQ01116399 GDIQ01098201 GDIP01135555 GDIQ01069465 GDIQ01054514 GDIQ01054513 JAL68159.1 JAN40224.1 GDIQ01017741 JAN76996.1 LRGB01002190 KZS08489.1 CM000160 EDW99278.1 CRL07381.1 AJWK01001387 APCN01004483 APCN01004484 HACA01029357 CDW46718.1 GL732645 EFX68939.1 CH933806 EDW15420.1 JRES01000409 KNC31499.1 KA649819 KA649831 AFP64448.1 CCAG010012940 JXJN01004056
KQ461187 KPJ07358.1 KP027422 AJL99442.1 NWSH01000041 PCG80338.1 BABH01007386 BABH01007387 AGBW02011913 OWR45931.1 GALX01003125 JAB65341.1 GECZ01023669 JAS46100.1 GEZM01013441 GEZM01013440 GEZM01013439 GEZM01013438 GEZM01013437 GEZM01013436 GEZM01013435 GEZM01013434 JAV92550.1 NEVH01026401 PNF14195.1 KQ971307 EFA11544.2 GBBI01001802 JAC16910.1 GECZ01032403 GECZ01006964 JAS37366.1 JAS62805.1 GECU01019070 GECU01016580 GECU01011865 JAS88636.1 JAS91126.1 JAS95841.1 GEBQ01014715 JAT25262.1 GBHO01019003 GBHO01019002 GDHC01014969 GDHC01014142 JAG24601.1 JAG24602.1 JAQ03660.1 JAQ04487.1 GDKW01002630 JAI53965.1 PNF14194.1 GECZ01021874 GECZ01013303 JAS47895.1 JAS56466.1 GECL01000821 JAP05303.1 GFTR01006110 JAW10316.1 KK852415 KDR24393.1 KU160500 AME17864.1 MG020525 AUC64081.1 GBHO01019001 GBHO01013057 JAG24603.1 JAG30547.1 GFDF01006447 JAV07637.1 GANP01010138 JAB74330.1 JH431704 GDRN01039794 GDRN01039793 JAI67184.1 GALA01001155 JAA93697.1 GFDF01006372 JAV07712.1 GGFL01004351 MBW68529.1 GGFM01003020 MBW23771.1 GGFM01003025 MBW23776.1 GGLE01006598 MBY10724.1 KQ435007 KZC13665.1 LNIX01000002 OXA59648.1 QOIP01000007 RLU20964.1 GGFK01002208 MBW35529.1 GL452201 EFN77795.1 DS232757 EDS45452.1 KK107238 EZA54816.1 GFDL01012807 JAV22238.1 GFWV01008619 MAA33348.1 AAAB01008966 BK000997 DAA00889.1 EAA13078.1 CVRI01000070 CRL07380.1 KQ976697 KYM77542.1 GANO01000322 JAB59549.1 GBBM01006544 JAC28874.1 JO841144 AEO32761.1 KQ977231 KYN04721.1 GBYB01009714 JAG79481.1 ADTU01020500 ADTU01020501 ADTU01020502 ADTU01020503 KQ978577 KYN30051.1 GL888172 EGI66122.1 GBBK01002492 JAC21990.1 KQ982727 KYQ51603.1 KQ981676 KYN38147.1 GL442363 EFN63489.1 JR037128 AEY57596.1 GEDV01003769 JAP84788.1 GFPF01006337 MAA17483.1 KZ288193 PBC34135.1 KQ435701 KOX80419.1 CH477828 EAT35854.1 CAEY01001996 CAEY01000418 GGFM01003077 MBW23828.1 ATLV01022194 KE525329 KFB47230.1 AXCN02001120 GFXV01004698 MBW16503.1 GGMR01003646 MBY16265.1 ABLF02027074 GFJQ02006144 JAW00826.1 GDIQ01185918 GDIQ01133752 GDIQ01116399 GDIQ01098201 GDIP01135555 GDIQ01069465 GDIQ01054514 GDIQ01054513 JAL68159.1 JAN40224.1 GDIQ01017741 JAN76996.1 LRGB01002190 KZS08489.1 CM000160 EDW99278.1 CRL07381.1 AJWK01001387 APCN01004483 APCN01004484 HACA01029357 CDW46718.1 GL732645 EFX68939.1 CH933806 EDW15420.1 JRES01000409 KNC31499.1 KA649819 KA649831 AFP64448.1 CCAG010012940 JXJN01004056
Proteomes
UP000053268
UP000053240
UP000218220
UP000005204
UP000007151
UP000235965
+ More
UP000007266 UP000027135 UP000192223 UP000076502 UP000198287 UP000279307 UP000008237 UP000002320 UP000053097 UP000076408 UP000007062 UP000183832 UP000078540 UP000078542 UP000005205 UP000078492 UP000007755 UP000075809 UP000078541 UP000000311 UP000242457 UP000005203 UP000053105 UP000069272 UP000008820 UP000015104 UP000030765 UP000075886 UP000075885 UP000075882 UP000007819 UP000075900 UP000076858 UP000002282 UP000092461 UP000075840 UP000000305 UP000009192 UP000037069 UP000092445 UP000095301 UP000092444 UP000092460
UP000007266 UP000027135 UP000192223 UP000076502 UP000198287 UP000279307 UP000008237 UP000002320 UP000053097 UP000076408 UP000007062 UP000183832 UP000078540 UP000078542 UP000005205 UP000078492 UP000007755 UP000075809 UP000078541 UP000000311 UP000242457 UP000005203 UP000053105 UP000069272 UP000008820 UP000015104 UP000030765 UP000075886 UP000075885 UP000075882 UP000007819 UP000075900 UP000076858 UP000002282 UP000092461 UP000075840 UP000000305 UP000009192 UP000037069 UP000092445 UP000095301 UP000092444 UP000092460
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
M1Q038
A0A2H1VRW2
A0A194PQ77
A0A0N1ID58
A0A0C5B7N5
A0A2A4K8U9
+ More
H9J264 A0A212EWT8 V5I9D5 A0A1B6F7C9 A0A1Y1N6J5 A0A2J7PCY8 D6W6M4 A0A023F6Q2 A0A1B6EHG1 A0A1B6IW07 A0A1B6LNF2 A0A0A9XV02 A0A0P4VWJ9 A0A2J7PCY3 A0A1B6G1Z7 A0A0V0GBD4 A0A224XCZ1 A0A067RMV7 A0A125S9L8 A0A2H4WAL5 A0A1W4XPX9 A0A0A9Y0J1 A0A1L8DMN7 V5GVA6 T1IZ20 A0A0N7ZDG5 T1E2L5 A0A1L8DMN8 A0A2M4CTC7 A0A2M3Z5H1 A0A2M3Z5I6 A0A2R5LMJ0 A0A154PP49 A0A226ER22 A0A3L8DKJ2 A0A2M4A446 E2C2N9 B0XDH4 A0A026WGJ5 A0A182Y727 A0A1Q3F422 A0A293LZZ7 A0A1S4H7B9 Q66GU5 Q7Q2V9 A0A1J1J6H2 A0A195AZS5 U5EP45 A0A023G4Y8 G3MGZ3 A0A195CVL7 A0A0C9R0P7 A0A158NMH5 A0A151JSD1 F4WI29 A0A023FMD3 A0A151WUS5 A0A151JVY0 E2ASV7 V9IAX1 A0A131Z1I8 A0A224YT92 A0A2A3EQU3 A0A087ZUT4 A0A0M9ACM6 A0A182FPL5 Q16NE5 T1KCR5 T1KRS3 A0A2M3Z5U6 A0A084WAI6 A0A182Q7L7 A0A182PGH6 A0A182KK83 A0A2H8TQN5 A0A2S2NGI2 X1X3E0 A0A1Z5L0B8 A0A0P6FT56 A0A182RVF8 A0A0P6I3D5 A0A164RB02 B4PP42 A0A1J1J4N9 A0A1B0C8T4 A0A2C9GQ18 A0A0K2V853 E9HH60 B4K866 A0A0L0CJ44 A0A1A9Z4B4 T1PM66 A0A1B0FL93 A0A1B0AV75
H9J264 A0A212EWT8 V5I9D5 A0A1B6F7C9 A0A1Y1N6J5 A0A2J7PCY8 D6W6M4 A0A023F6Q2 A0A1B6EHG1 A0A1B6IW07 A0A1B6LNF2 A0A0A9XV02 A0A0P4VWJ9 A0A2J7PCY3 A0A1B6G1Z7 A0A0V0GBD4 A0A224XCZ1 A0A067RMV7 A0A125S9L8 A0A2H4WAL5 A0A1W4XPX9 A0A0A9Y0J1 A0A1L8DMN7 V5GVA6 T1IZ20 A0A0N7ZDG5 T1E2L5 A0A1L8DMN8 A0A2M4CTC7 A0A2M3Z5H1 A0A2M3Z5I6 A0A2R5LMJ0 A0A154PP49 A0A226ER22 A0A3L8DKJ2 A0A2M4A446 E2C2N9 B0XDH4 A0A026WGJ5 A0A182Y727 A0A1Q3F422 A0A293LZZ7 A0A1S4H7B9 Q66GU5 Q7Q2V9 A0A1J1J6H2 A0A195AZS5 U5EP45 A0A023G4Y8 G3MGZ3 A0A195CVL7 A0A0C9R0P7 A0A158NMH5 A0A151JSD1 F4WI29 A0A023FMD3 A0A151WUS5 A0A151JVY0 E2ASV7 V9IAX1 A0A131Z1I8 A0A224YT92 A0A2A3EQU3 A0A087ZUT4 A0A0M9ACM6 A0A182FPL5 Q16NE5 T1KCR5 T1KRS3 A0A2M3Z5U6 A0A084WAI6 A0A182Q7L7 A0A182PGH6 A0A182KK83 A0A2H8TQN5 A0A2S2NGI2 X1X3E0 A0A1Z5L0B8 A0A0P6FT56 A0A182RVF8 A0A0P6I3D5 A0A164RB02 B4PP42 A0A1J1J4N9 A0A1B0C8T4 A0A2C9GQ18 A0A0K2V853 E9HH60 B4K866 A0A0L0CJ44 A0A1A9Z4B4 T1PM66 A0A1B0FL93 A0A1B0AV75
PDB
2WTR
E-value=1.08223e-133,
Score=1221
Ontologies
GO
PANTHER
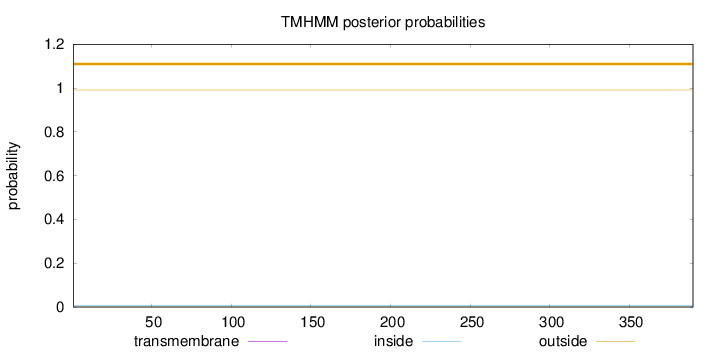
Topology
Length:
390
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00037
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00849
outside
1 - 390
Population Genetic Test Statistics
Pi
144.674558
Theta
135.985465
Tajima's D
0
CLR
109.512959
CSRT
0.371881405929704
Interpretation
Uncertain
