Gene
KWMTBOMO03015
Pre Gene Modal
BGIBMGA003594
Annotation
PREDICTED:_kinesin-like_protein_KIF21B_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.352
Sequence
CDS
ATGGAACCGTGTAAAGAGCCGGCAAGCACGCCGCCCGCCTCTCCGGGGGCCACGAGACGAGCCAGGGATGACGACGTGTTCTTGCGACTAGCGGGCGCCGGACACGATAACTCGCCCCAAGGAATCGTCAAAGAACTTGCAATCAAACGCGTTATCGGTGGAAACACGAGAGCGTGGCTGCAGTGCACACACGTGGCGGAGGGACACGCGGGCGCGGCTCTGGCCCTGGCCGCCACAACACACGCTGTGTACAGCGGCGGCGTAGATCGCACGGTCCGTGGCTGGGACCTGTCCGTGGGCAGCGAGGCGTGGCGCGGCTGGTGCGGCGGCGCGGTGTCGTCGCTGGCGGCCGGCACCGCCGCGGACGGGGGCAGGCTGCTCCTCGCCGCCGCCGGTCCCGCCGTCAGGCTCTTCGACACCCGGACTAGGACACCGCTCACTACGCTCTGGTCGTCGGGCGCGACCGGTCCGTGTCCGGCGGGGCGCGGCGCGGCCGGCGAGGTGCCGGTGACGGCGCTGCGGCTGTGCGCGCCTCACACGCTGTACACCGCCGCGGGGGACAAGCTGCGTCTGTGGGACCTGCGCATGTTGGAGTGCGTGTACAAGATATGGTCGGGCCACGCGGCCGCCGTCATGTGTCTGGCGGCCGCCGATAACTTGCTCGTCACCGGCTCGAAGGACCATTATGTCAGAGCCATGGAGATAAACGCACAAGACTCAGGAGGTTGGGAGACGAGCAACCGGCGCCTGCTGGAGCCCCCGCACTACGACGGCGTGCAGGCGCTCGCCCTGCGCGGCCACGTGCTGTACAGCGCCTCGCGGGACACCAGCCTCAAGCGCTGGAACTTGCACGACAACTCGCTCACCCATAGCGTGATGAACGCGCACAAGGGCTGGGTGACGGGCGTGTGCGTGCTGGGCGGCGCGGAGGAGGGCGCGGTGGCGAGCTGCGGGCGCGAGGGCGCCGTGCGCGTGTGGAGCGCCGACCTGCGCGCCGTGACGTCACCCGCCGCGCTCTCCGACGCCGTGCACGCGCTCGACGCGCACCCCGACCCGCACCGCCGGACGCTCTACACCGCCTCCAACGGCGGCGAATTGTCGTAG
Protein
MEPCKEPASTPPASPGATRRARDDDVFLRLAGAGHDNSPQGIVKELAIKRVIGGNTRAWLQCTHVAEGHAGAALALAATTHAVYSGGVDRTVRGWDLSVGSEAWRGWCGGAVSSLAAGTAADGGRLLLAAAGPAVRLFDTRTRTPLTTLWSSGATGPCPAGRGAAGEVPVTALRLCAPHTLYTAAGDKLRLWDLRMLECVYKIWSGHAAAVMCLAAADNLLVTGSKDHYVRAMEINAQDSGGWETSNRRLLEPPHYDGVQALALRGHVLYSASRDTSLKRWNLHDNSLTHSVMNAHKGWVTGVCVLGGAEEGAVASCGREGAVRVWSADLRAVTSPAALSDAVHALDAHPDPHRRTLYTASNGGELS
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9J257
A0A2A4K9C6
A0A212EWS3
A0A194PVG0
W4VRN1
B0W1P9
+ More
A0A182F3X7 A0A182XZK5 Q7PN43 A0A1Q3FTU7 A0A182MUL0 W5J9J8 A0A1Q3FU77 A0A1Q3FUB0 A0A1S4GWC8 A0A182JV59 A0A182THB3 A0A1B6HC47 A0A1B6ISA5 A0A182V5F7 A0A182MZD8 A0A182IEI6 A0A182P0F1 A0A1B6MHN2 A0A1B6L5I8 A0A182X1K6 A0A1B6LXU8 A0A182JDN6 A0A1B6D0V6 A0A1B6CPU2 Q17KL6 A0A182W7T1 A0A224XRX5 A0A1J1ILK2 A0A1B0DNE1 A0A2S2QRQ4 A0A2H8TLV6 A0A023ENV3 J9K1M7 A0A2S2NQX3 E0VQK1 A0A2A3EKG5 A0A026WE30 A0A088A5V3 E2BPS6 A0A0K8T836 A0A151K056 A0A195BBF3 A0A2T7PKG5 V5HJ08 A0A151IVG4 V5IJV2 A0A182RVM3 A0A087SXI4 K7IQH0 F4WE20 A0A1V9XUW5 A0A151IN70 E2A764 A0A158NUS8 A0A2R7VS35 A0A151WNS1 A0A0K2SV43 A0A2P6L8L0 T1HW35 A0A2R5LKP4 A0A0M8ZWE4 A0A0A9XTB7 A0A154P5A3 W4YK07 A0A224YXF4 A0A0A9XQ40 A0A0A9XXR5 A0A093G812 A0A091SEX5 L7LX05 A0A2P2ICN6 A0A093I342 A0A226EJN6 A0A1D5PRV0 A0A3B3TMP1 A0A1D5PVG4 A0A091LIT2 A0A2G8KEY1 A0A0L7R1V1 A0A1S3WM71 A0A1S3WMT3 A0A1S3WM43 A0A0A0A8P7 A0A3B3TJW3 A0A091FSL7
A0A182F3X7 A0A182XZK5 Q7PN43 A0A1Q3FTU7 A0A182MUL0 W5J9J8 A0A1Q3FU77 A0A1Q3FUB0 A0A1S4GWC8 A0A182JV59 A0A182THB3 A0A1B6HC47 A0A1B6ISA5 A0A182V5F7 A0A182MZD8 A0A182IEI6 A0A182P0F1 A0A1B6MHN2 A0A1B6L5I8 A0A182X1K6 A0A1B6LXU8 A0A182JDN6 A0A1B6D0V6 A0A1B6CPU2 Q17KL6 A0A182W7T1 A0A224XRX5 A0A1J1ILK2 A0A1B0DNE1 A0A2S2QRQ4 A0A2H8TLV6 A0A023ENV3 J9K1M7 A0A2S2NQX3 E0VQK1 A0A2A3EKG5 A0A026WE30 A0A088A5V3 E2BPS6 A0A0K8T836 A0A151K056 A0A195BBF3 A0A2T7PKG5 V5HJ08 A0A151IVG4 V5IJV2 A0A182RVM3 A0A087SXI4 K7IQH0 F4WE20 A0A1V9XUW5 A0A151IN70 E2A764 A0A158NUS8 A0A2R7VS35 A0A151WNS1 A0A0K2SV43 A0A2P6L8L0 T1HW35 A0A2R5LKP4 A0A0M8ZWE4 A0A0A9XTB7 A0A154P5A3 W4YK07 A0A224YXF4 A0A0A9XQ40 A0A0A9XXR5 A0A093G812 A0A091SEX5 L7LX05 A0A2P2ICN6 A0A093I342 A0A226EJN6 A0A1D5PRV0 A0A3B3TMP1 A0A1D5PVG4 A0A091LIT2 A0A2G8KEY1 A0A0L7R1V1 A0A1S3WM71 A0A1S3WMT3 A0A1S3WM43 A0A0A0A8P7 A0A3B3TJW3 A0A091FSL7
Pubmed
EMBL
BABH01007392
BABH01007393
BABH01007394
BABH01007395
BABH01007396
NWSH01000041
+ More
PCG80340.1 AGBW02011913 OWR45933.1 KQ459597 KPI95115.1 GANO01001312 JAB58559.1 DS231823 EDS26600.1 AAAB01008964 EAA12817.4 GFDL01004031 JAV31014.1 AXCM01003209 ADMH02001984 ETN60083.1 GFDL01004033 JAV31012.1 GFDL01004032 JAV31013.1 GECU01035455 GECU01030997 GECU01023851 GECU01020998 GECU01012222 JAS72251.1 JAS76709.1 JAS83855.1 JAS86708.1 JAS95484.1 GECU01031725 GECU01028530 GECU01021358 GECU01017895 JAS75981.1 JAS79176.1 JAS86348.1 JAS89811.1 APCN01005204 GEBQ01004551 JAT35426.1 GEBQ01020985 JAT18992.1 GEBQ01011465 JAT28512.1 GEDC01018011 JAS19287.1 GEDC01021907 JAS15391.1 CH477223 EAT47210.1 GFTR01005236 JAW11190.1 CVRI01000054 CRL00444.1 AJVK01007518 AJVK01007519 AJVK01007520 GGMS01011208 MBY80411.1 GFXV01003329 MBW15134.1 GAPW01002476 JAC11122.1 ABLF02035294 GGMR01006733 MBY19352.1 DS235430 EEB15657.1 KZ288229 PBC31686.1 KK107261 QOIP01000012 EZA53931.1 RLU16343.1 GL449658 EFN82277.1 GBRD01004114 JAG61707.1 KQ981296 KYN43098.1 KQ976532 KYM81545.1 PZQS01000003 PVD33894.1 GANP01000793 JAB83675.1 KQ980926 KYN11395.1 GANP01000792 JAB83676.1 KK112404 KFM57573.1 GL888102 EGI67458.1 MNPL01003729 OQR77297.1 KQ976959 KYN06791.1 GL437267 EFN70720.1 ADTU01026692 KK854024 PTY09651.1 KQ982905 KYQ49493.1 HACA01000093 HACA01000094 CDW17454.1 MWRG01000964 PRD34923.1 ACPB03022165 GGLE01005957 MBY10083.1 KQ435848 KOX70983.1 GBHO01021526 JAG22078.1 KQ434809 KZC06514.1 AAGJ04050249 AAGJ04050250 AAGJ04050251 AAGJ04050252 AAGJ04050253 AAGJ04050254 AAGJ04050255 GFPF01009263 MAA20409.1 GBHO01021535 JAG22069.1 GBHO01021534 JAG22070.1 KL215028 KFV62987.1 KK949155 KFQ56903.1 GACK01009596 JAA55438.1 IACF01006186 LAB71769.1 KK586053 KFV97084.1 LNIX01000003 OXA57214.1 AC192335 KL322830 KFP55256.1 MRZV01000639 PIK46525.1 KQ414667 KOC64824.1 KL871087 KGL90322.1 KL447430 KFO73505.1
PCG80340.1 AGBW02011913 OWR45933.1 KQ459597 KPI95115.1 GANO01001312 JAB58559.1 DS231823 EDS26600.1 AAAB01008964 EAA12817.4 GFDL01004031 JAV31014.1 AXCM01003209 ADMH02001984 ETN60083.1 GFDL01004033 JAV31012.1 GFDL01004032 JAV31013.1 GECU01035455 GECU01030997 GECU01023851 GECU01020998 GECU01012222 JAS72251.1 JAS76709.1 JAS83855.1 JAS86708.1 JAS95484.1 GECU01031725 GECU01028530 GECU01021358 GECU01017895 JAS75981.1 JAS79176.1 JAS86348.1 JAS89811.1 APCN01005204 GEBQ01004551 JAT35426.1 GEBQ01020985 JAT18992.1 GEBQ01011465 JAT28512.1 GEDC01018011 JAS19287.1 GEDC01021907 JAS15391.1 CH477223 EAT47210.1 GFTR01005236 JAW11190.1 CVRI01000054 CRL00444.1 AJVK01007518 AJVK01007519 AJVK01007520 GGMS01011208 MBY80411.1 GFXV01003329 MBW15134.1 GAPW01002476 JAC11122.1 ABLF02035294 GGMR01006733 MBY19352.1 DS235430 EEB15657.1 KZ288229 PBC31686.1 KK107261 QOIP01000012 EZA53931.1 RLU16343.1 GL449658 EFN82277.1 GBRD01004114 JAG61707.1 KQ981296 KYN43098.1 KQ976532 KYM81545.1 PZQS01000003 PVD33894.1 GANP01000793 JAB83675.1 KQ980926 KYN11395.1 GANP01000792 JAB83676.1 KK112404 KFM57573.1 GL888102 EGI67458.1 MNPL01003729 OQR77297.1 KQ976959 KYN06791.1 GL437267 EFN70720.1 ADTU01026692 KK854024 PTY09651.1 KQ982905 KYQ49493.1 HACA01000093 HACA01000094 CDW17454.1 MWRG01000964 PRD34923.1 ACPB03022165 GGLE01005957 MBY10083.1 KQ435848 KOX70983.1 GBHO01021526 JAG22078.1 KQ434809 KZC06514.1 AAGJ04050249 AAGJ04050250 AAGJ04050251 AAGJ04050252 AAGJ04050253 AAGJ04050254 AAGJ04050255 GFPF01009263 MAA20409.1 GBHO01021535 JAG22069.1 GBHO01021534 JAG22070.1 KL215028 KFV62987.1 KK949155 KFQ56903.1 GACK01009596 JAA55438.1 IACF01006186 LAB71769.1 KK586053 KFV97084.1 LNIX01000003 OXA57214.1 AC192335 KL322830 KFP55256.1 MRZV01000639 PIK46525.1 KQ414667 KOC64824.1 KL871087 KGL90322.1 KL447430 KFO73505.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000002320
UP000069272
+ More
UP000076408 UP000007062 UP000075883 UP000000673 UP000075881 UP000075902 UP000075903 UP000075884 UP000075840 UP000075885 UP000076407 UP000075880 UP000008820 UP000075920 UP000183832 UP000092462 UP000007819 UP000009046 UP000242457 UP000053097 UP000279307 UP000005203 UP000008237 UP000078541 UP000078540 UP000245119 UP000078492 UP000075900 UP000054359 UP000002358 UP000007755 UP000192247 UP000078542 UP000000311 UP000005205 UP000075809 UP000015103 UP000053105 UP000076502 UP000007110 UP000053875 UP000198287 UP000000539 UP000261500 UP000230750 UP000053825 UP000079721 UP000053858 UP000053760
UP000076408 UP000007062 UP000075883 UP000000673 UP000075881 UP000075902 UP000075903 UP000075884 UP000075840 UP000075885 UP000076407 UP000075880 UP000008820 UP000075920 UP000183832 UP000092462 UP000007819 UP000009046 UP000242457 UP000053097 UP000279307 UP000005203 UP000008237 UP000078541 UP000078540 UP000245119 UP000078492 UP000075900 UP000054359 UP000002358 UP000007755 UP000192247 UP000078542 UP000000311 UP000005205 UP000075809 UP000015103 UP000053105 UP000076502 UP000007110 UP000053875 UP000198287 UP000000539 UP000261500 UP000230750 UP000053825 UP000079721 UP000053858 UP000053760
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR036322 WD40_repeat_dom_sf
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR020472 G-protein_beta_WD-40_rep
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR036322 WD40_repeat_dom_sf
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR020472 G-protein_beta_WD-40_rep
Gene 3D
ProteinModelPortal
H9J257
A0A2A4K9C6
A0A212EWS3
A0A194PVG0
W4VRN1
B0W1P9
+ More
A0A182F3X7 A0A182XZK5 Q7PN43 A0A1Q3FTU7 A0A182MUL0 W5J9J8 A0A1Q3FU77 A0A1Q3FUB0 A0A1S4GWC8 A0A182JV59 A0A182THB3 A0A1B6HC47 A0A1B6ISA5 A0A182V5F7 A0A182MZD8 A0A182IEI6 A0A182P0F1 A0A1B6MHN2 A0A1B6L5I8 A0A182X1K6 A0A1B6LXU8 A0A182JDN6 A0A1B6D0V6 A0A1B6CPU2 Q17KL6 A0A182W7T1 A0A224XRX5 A0A1J1ILK2 A0A1B0DNE1 A0A2S2QRQ4 A0A2H8TLV6 A0A023ENV3 J9K1M7 A0A2S2NQX3 E0VQK1 A0A2A3EKG5 A0A026WE30 A0A088A5V3 E2BPS6 A0A0K8T836 A0A151K056 A0A195BBF3 A0A2T7PKG5 V5HJ08 A0A151IVG4 V5IJV2 A0A182RVM3 A0A087SXI4 K7IQH0 F4WE20 A0A1V9XUW5 A0A151IN70 E2A764 A0A158NUS8 A0A2R7VS35 A0A151WNS1 A0A0K2SV43 A0A2P6L8L0 T1HW35 A0A2R5LKP4 A0A0M8ZWE4 A0A0A9XTB7 A0A154P5A3 W4YK07 A0A224YXF4 A0A0A9XQ40 A0A0A9XXR5 A0A093G812 A0A091SEX5 L7LX05 A0A2P2ICN6 A0A093I342 A0A226EJN6 A0A1D5PRV0 A0A3B3TMP1 A0A1D5PVG4 A0A091LIT2 A0A2G8KEY1 A0A0L7R1V1 A0A1S3WM71 A0A1S3WMT3 A0A1S3WM43 A0A0A0A8P7 A0A3B3TJW3 A0A091FSL7
A0A182F3X7 A0A182XZK5 Q7PN43 A0A1Q3FTU7 A0A182MUL0 W5J9J8 A0A1Q3FU77 A0A1Q3FUB0 A0A1S4GWC8 A0A182JV59 A0A182THB3 A0A1B6HC47 A0A1B6ISA5 A0A182V5F7 A0A182MZD8 A0A182IEI6 A0A182P0F1 A0A1B6MHN2 A0A1B6L5I8 A0A182X1K6 A0A1B6LXU8 A0A182JDN6 A0A1B6D0V6 A0A1B6CPU2 Q17KL6 A0A182W7T1 A0A224XRX5 A0A1J1ILK2 A0A1B0DNE1 A0A2S2QRQ4 A0A2H8TLV6 A0A023ENV3 J9K1M7 A0A2S2NQX3 E0VQK1 A0A2A3EKG5 A0A026WE30 A0A088A5V3 E2BPS6 A0A0K8T836 A0A151K056 A0A195BBF3 A0A2T7PKG5 V5HJ08 A0A151IVG4 V5IJV2 A0A182RVM3 A0A087SXI4 K7IQH0 F4WE20 A0A1V9XUW5 A0A151IN70 E2A764 A0A158NUS8 A0A2R7VS35 A0A151WNS1 A0A0K2SV43 A0A2P6L8L0 T1HW35 A0A2R5LKP4 A0A0M8ZWE4 A0A0A9XTB7 A0A154P5A3 W4YK07 A0A224YXF4 A0A0A9XQ40 A0A0A9XXR5 A0A093G812 A0A091SEX5 L7LX05 A0A2P2ICN6 A0A093I342 A0A226EJN6 A0A1D5PRV0 A0A3B3TMP1 A0A1D5PVG4 A0A091LIT2 A0A2G8KEY1 A0A0L7R1V1 A0A1S3WM71 A0A1S3WMT3 A0A1S3WM43 A0A0A0A8P7 A0A3B3TJW3 A0A091FSL7
PDB
4JXM
E-value=7.26136e-06,
Score=118
Ontologies
GO
PANTHER
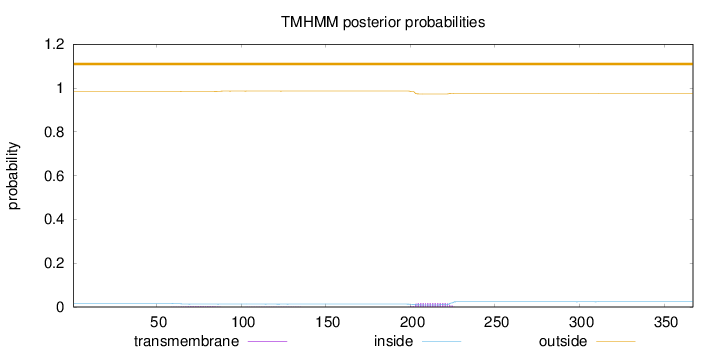
Topology
Length:
367
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.457890000000001
Exp number, first 60 AAs:
0.00058
Total prob of N-in:
0.01534
outside
1 - 367
Population Genetic Test Statistics
Pi
314.464549
Theta
162.421373
Tajima's D
2.236945
CLR
0.291468
CSRT
0.91730413479326
Interpretation
Uncertain
