Gene
KWMTBOMO03014
Pre Gene Modal
BGIBMGA003594
Annotation
PREDICTED:_kinesin-like_protein_KIF21B_isoform_X4_[Bombyx_mori]
Full name
Kinesin-like protein
Location in the cell
Nuclear Reliability : 3.346
Sequence
CDS
ATGGCCAGTGAGGAATCCAGCGTAAGAGTTGCTGTGAGGATTCGTCCACAAACACCAGCCGAGATTGTGGAGGGATGCGGTATATGCGCGAGAGCAGGTGGTCCAGCGGGCGAGGGTGGGGTGGCTTTGGGCACCGAAAGAGCCTTCACCTTCGATTACGCCTTCGAACCTTCAGCTTCGCAACAGGAGCTGTACGAAACCTGCGTCAAGAAACTGGTGGAAGCCGCTCTTGATGGATACAACGCTACCGTGCTGGCATATGGACAGACCGGCTCCGGTAAGACATACACGATGGGCAGCGGCTGGGAAGGCGAGGGAGACGGTGATGAGGACAGGCGCGGCATCATCCCTCGGGCGATCCGGGATCTGTTCGCGGGCGCGGACGCTCGCGCTGATGAGGCACGCACACAGGGTCAGCTGCCCCCCGAGTTCTCCGTCCAGGCTCAGTTCATTGAACTGTACAATGAGGATATCGTTGATTTATTGGATCCGGCCAAAGATCCATTTTCTAAGGGAGCGCTGAGAATAACAGAGGACGGGTGCGGCGGGGTGCGTGTTGTGGGCGCTTCGATGCGGGCGGTCCGGGGCGCCAGAGAAGCCCTGGGCGCTCTGAGGGCCGGCGCCCTTGCTCGAACAACGGCGGCCACCAACATGAACTCCTCATCGTCGCGGTCGCACGCAGTCTTCACGTTGCTGTTACGTCAACGGAGGCTCGCCGCGGACTTACCTGACGGCATAGAAAGGGACGCGGAGTCAGATTCTCCCGAACAATATGAAACTTTAACTGCCAAGTTCCATTTCGTCGATCTAGCTGGATCTGAACGCTTGAAGAGGACTGGTGCGACGGGAGAGCGCGCTCGCGAGGGCATATCGATAAACTGTGGACTACTGGCGCTGGGCAACGTCATATCCGCACTCGGAGACAAGTCGAGGAAGGCGCTGCACGTGCCCTACAGAGATTCTAAGCTGACGAGACTGTTGCAGGATTCGTTGGGCGGTAACAGCAACACAGTGATGATAGCATGCGTGTCGCCGAGCGACCGCGACTTCATGGAGACCTTGAACACGTTGAAATACGCGAACCGAGCTCGGAACATCAAGAACCGTTGCGTCGTCAACCAGGACCTCACCTCCAGGACCATACACCAGCTGCGCCAGGAGGTCGCGCGGCTGCAACTCGAACTAGCGGAATACAAACAGGGAAAACGCGTAATATCAGAAAACGGCGAGGAGGGTTGGAGCGACGTCGTCCAAGAGAACGCTATACTGAATGGAGAAGTTGAAGCACTTAGACGTCGCGTCAAAGCCATGCAGGGCACCATAGATCAGCTGTCGGCCAGGAACAGCGAACTGATGGCTGAGAAGACGCTGAGTAACTGGATTCCGAAGGACGGCAGCCCTGACACCAACGATTGCTCCCTCACCACTCTCGTCCAGGGCTACGTGAGCGAGATAGAGAGTCTACGAGCCCAGCTGATGGAGACGACGGCGATGTACGAGGCGAGCCGGCGCCGCGAGCAGATGACGTCACGCACCCGCCACGAGTCCGCGCACGACCCCGCCGCCGTCATCGACGAGGCCAAGCGGGAACTGTACAAGGAAAAAGAAATTCTGGCGCGGAGCATGGGCGAACTGGAGTTCCACCGCAAACTGAACATGAGCGACAGTGTGATGAGCAGCGTGGACGATCGGCCCATAGGAGAGAGGGAAAGGGCTGAGGGTGAGAGTGCCGATGACTCCGATCCTTCGGACGGGGAAGCCGATGGAGATGGCACTGATTCCGACGAAGATTCTGAGACTAAAGGACAACGTGTACTATCAGCGCAACTGGTAGCCCTCAGCGAGGACATCGATACGAAAGCTCGTCTTATCGAGCAACTAGAGATGTCCCAGCGGCGGCTCACCGCCCTCCGGACCCACTACGAACAGAGGCTCGACACGCTGCATCATCAGATCAAGGCCACCAACGACGAGAAAGATAAAGTCTTGGCTTCTTTAGCCGCGCAAGCGTCTCCGGCGAGCGAGAAGGTGAAGCGTGTCCGCGAGGAGTACGAGCGGCGCGCGGCGGGCATGGCGCGCGAGCTGCGGCGCCTGCACGCCGCGCACCGCGACCACGCGCGGCTGCAGCGGGCGCAGGCGCACACCGCCGTGCAGATCTCCACGCTCAGGAGCGAACTGCAGAACATGAAGAGGGATAAGGTAAAATTAGTCCAACAAATGCGTGCTGAGGCCAAACGACACGCTCAAGCAGAAGCTGTTAGAGCTAAAGAGGTCGCTCAACTGCGCAAGGAATCCCGTAAAAATGCAAATCTCATCAGATCTCTGGAGGCTGAAACGAAACTCAAAGAACAGGTATTGAAACGGAAACAAGAGGAAGTGTCTCTATTGAGACGCGGACATCGCGATAAGTTAAGCAATCGAGCCGCCGGGCGACTGTACGGAGGGGTCCGCGGTCGCAGCCGCAAGTTGGTCCGCGAGAGCTGGGCGCGGCTGGAGCGGTGGGTCGCCCGCACCAGCGCCGCCCGCCTCACGCTGTGGGAGCTGGAGGGCGCGCTGGACCGCCTGCTGCGGCAGCGCGCGGCGGCGCAGCGGGCGGCGCACGACGGCGACGCCGCCTCGCTGCACGCCTACCTGCGCGACGCCATCGCCGACACGCAGGCGCAGATTATGCAGATCGAGGAAGAAAACGACGAGAACGAACTGTCCCGGATACTGGAGCAGGTGGACAGCGCGGAGGCGGGCCGGTACGCGCTGGAGCGGCTGGCGGCGTGCGCGCTGCAGCACGCGCAGGACGCCGCCAGGAACCTCAAGCTGCTCAACGACACACGCGCGCATCTCAAAGAATTGGAGGAGAGCCAAGAGCGAGCCGCCAGCGCTCTCCGGGCGGCGGAGGAACAGAACCTCACGAGCTGGAGCTGCGGCACCGCCCTGGCTCACCTGCTGGCGCACGTCTCCTCCGGTACATCCACCAGATCGGTCTCGCCGGTCGATAGTTCAATATTGGAATCGAAGCCCAACACGAACGGCACTAGGGAGACGGCGACGGCGCCCACCTCGCCGCCCGACGACAACGTCAATTCGCCCTTCCAAAGAAATACAGTACGTCGCGGCTCGGTGCGTCTCCGCGACCTGGGCGTCATCGCCAGAGAGGACGGGGACGACCCTATGACGCAGTCGCTGGTGGAGCCCACCTCGCCCCGCCCCGCGCCCCTCAGCCGCGTGCCCAGCGCCCCCGGCAGTCTGAGATACCACAACGTGATTTTCACTAATGTTAGAATATGCTAA
Protein
MASEESSVRVAVRIRPQTPAEIVEGCGICARAGGPAGEGGVALGTERAFTFDYAFEPSASQQELYETCVKKLVEAALDGYNATVLAYGQTGSGKTYTMGSGWEGEGDGDEDRRGIIPRAIRDLFAGADARADEARTQGQLPPEFSVQAQFIELYNEDIVDLLDPAKDPFSKGALRITEDGCGGVRVVGASMRAVRGAREALGALRAGALARTTAATNMNSSSSRSHAVFTLLLRQRRLAADLPDGIERDAESDSPEQYETLTAKFHFVDLAGSERLKRTGATGERAREGISINCGLLALGNVISALGDKSRKALHVPYRDSKLTRLLQDSLGGNSNTVMIACVSPSDRDFMETLNTLKYANRARNIKNRCVVNQDLTSRTIHQLRQEVARLQLELAEYKQGKRVISENGEEGWSDVVQENAILNGEVEALRRRVKAMQGTIDQLSARNSELMAEKTLSNWIPKDGSPDTNDCSLTTLVQGYVSEIESLRAQLMETTAMYEASRRREQMTSRTRHESAHDPAAVIDEAKRELYKEKEILARSMGELEFHRKLNMSDSVMSSVDDRPIGERERAEGESADDSDPSDGEADGDGTDSDEDSETKGQRVLSAQLVALSEDIDTKARLIEQLEMSQRRLTALRTHYEQRLDTLHHQIKATNDEKDKVLASLAAQASPASEKVKRVREEYERRAAGMARELRRLHAAHRDHARLQRAQAHTAVQISTLRSELQNMKRDKVKLVQQMRAEAKRHAQAEAVRAKEVAQLRKESRKNANLIRSLEAETKLKEQVLKRKQEEVSLLRRGHRDKLSNRAAGRLYGGVRGRSRKLVRESWARLERWVARTSAARLTLWELEGALDRLLRQRAAAQRAAHDGDAASLHAYLRDAIADTQAQIMQIEEENDENELSRILEQVDSAEAGRYALERLAACALQHAQDAARNLKLLNDTRAHLKELEESQERAASALRAAEEQNLTSWSCGTALAHLLAHVSSGTSTRSVSPVDSSILESKPNTNGTRETATAPTSPPDDNVNSPFQRNTVRRGSVRLRDLGVIAREDGDDPMTQSLVEPTSPRPAPLSRVPSAPGSLRYHNVIFTNVRIC
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9J257
A0A2A4K9C6
A0A0N1INJ2
A0A212EWS3
A0A194PVG0
A0A2H1VC82
+ More
A0A1B6EGH2 A0A1Y1MV48 A0A1W4X8E9 A0A1B6C896 A0A1B6CPU2 A0A1Y1MWA9 D6W864 F4WE20 A0A026WE30 A0A158NUS8 A0A195BBF3 E2BPS6 A0A1W4WXL2 A0A1B6EWN8 A0A1B6EI10 A0A2S2QRQ4 N6TG63 A0A2A3EKG5 A0A088A5V3 A0A151WNS1 A0A0L7R1V1 A0A151IN70 A0A2S2NQX3 J9K1M7 A0A151IVG4 A0A151K056 A0A1B0DNE0 E2A764 A0A0M8ZWE4 A0A131XJE8 A0A1A9Y5P3 A0A2K6FSD5 A0A2K6FSE8 A0A2K6FSD7 A0A2R9C2S8 H2R894 A0A131YVY1 A0A2K5RNK4 F6WVK7 A0A224YXF4 A0A2K5CNV3 L7LX05 A0A1A7W978 A0A1A8CTM6 A0A2I4C2U3 A0A3B4UDC4 A0A3B4UD73 A0A2I4C2T6 A0A3B3VKU7 A0A3P9NPL6 A0A146VH53 A0A146ZQU4
A0A1B6EGH2 A0A1Y1MV48 A0A1W4X8E9 A0A1B6C896 A0A1B6CPU2 A0A1Y1MWA9 D6W864 F4WE20 A0A026WE30 A0A158NUS8 A0A195BBF3 E2BPS6 A0A1W4WXL2 A0A1B6EWN8 A0A1B6EI10 A0A2S2QRQ4 N6TG63 A0A2A3EKG5 A0A088A5V3 A0A151WNS1 A0A0L7R1V1 A0A151IN70 A0A2S2NQX3 J9K1M7 A0A151IVG4 A0A151K056 A0A1B0DNE0 E2A764 A0A0M8ZWE4 A0A131XJE8 A0A1A9Y5P3 A0A2K6FSD5 A0A2K6FSE8 A0A2K6FSD7 A0A2R9C2S8 H2R894 A0A131YVY1 A0A2K5RNK4 F6WVK7 A0A224YXF4 A0A2K5CNV3 L7LX05 A0A1A7W978 A0A1A8CTM6 A0A2I4C2U3 A0A3B4UDC4 A0A3B4UD73 A0A2I4C2T6 A0A3B3VKU7 A0A3P9NPL6 A0A146VH53 A0A146ZQU4
Pubmed
EMBL
BABH01007392
BABH01007393
BABH01007394
BABH01007395
BABH01007396
NWSH01000041
+ More
PCG80340.1 KQ461187 KPJ07356.1 AGBW02011913 OWR45933.1 KQ459597 KPI95115.1 ODYU01001783 SOQ38459.1 GEDC01000283 JAS37015.1 GEZM01020070 JAV89419.1 GEDC01027580 JAS09718.1 GEDC01021907 JAS15391.1 GEZM01020071 JAV89418.1 KQ971307 EFA11192.2 GL888102 EGI67458.1 KK107261 QOIP01000012 EZA53931.1 RLU16343.1 ADTU01026692 KQ976532 KYM81545.1 GL449658 EFN82277.1 GECZ01027489 JAS42280.1 GECZ01032206 JAS37563.1 GGMS01011208 MBY80411.1 APGK01039035 KB740967 KB632399 ENN76758.1 ERL94755.1 KZ288229 PBC31686.1 KQ982905 KYQ49493.1 KQ414667 KOC64824.1 KQ976959 KYN06791.1 GGMR01006733 MBY19352.1 ABLF02035294 KQ980926 KYN11395.1 KQ981296 KYN43098.1 AJVK01007515 AJVK01007516 AJVK01007517 AJVK01007518 GL437267 EFN70720.1 KQ435848 KOX70983.1 GEFH01002920 JAP65661.1 AJFE02080412 AJFE02080413 AACZ04071406 GEDV01005845 JAP82712.1 JSUE03003052 JSUE03003053 GFPF01009263 MAA20409.1 GACK01009596 JAA55438.1 HADW01000860 SBP02260.1 HADZ01019158 SBP83099.1 GCES01069657 JAR16666.1 GCES01017633 JAR68690.1
PCG80340.1 KQ461187 KPJ07356.1 AGBW02011913 OWR45933.1 KQ459597 KPI95115.1 ODYU01001783 SOQ38459.1 GEDC01000283 JAS37015.1 GEZM01020070 JAV89419.1 GEDC01027580 JAS09718.1 GEDC01021907 JAS15391.1 GEZM01020071 JAV89418.1 KQ971307 EFA11192.2 GL888102 EGI67458.1 KK107261 QOIP01000012 EZA53931.1 RLU16343.1 ADTU01026692 KQ976532 KYM81545.1 GL449658 EFN82277.1 GECZ01027489 JAS42280.1 GECZ01032206 JAS37563.1 GGMS01011208 MBY80411.1 APGK01039035 KB740967 KB632399 ENN76758.1 ERL94755.1 KZ288229 PBC31686.1 KQ982905 KYQ49493.1 KQ414667 KOC64824.1 KQ976959 KYN06791.1 GGMR01006733 MBY19352.1 ABLF02035294 KQ980926 KYN11395.1 KQ981296 KYN43098.1 AJVK01007515 AJVK01007516 AJVK01007517 AJVK01007518 GL437267 EFN70720.1 KQ435848 KOX70983.1 GEFH01002920 JAP65661.1 AJFE02080412 AJFE02080413 AACZ04071406 GEDV01005845 JAP82712.1 JSUE03003052 JSUE03003053 GFPF01009263 MAA20409.1 GACK01009596 JAA55438.1 HADW01000860 SBP02260.1 HADZ01019158 SBP83099.1 GCES01069657 JAR16666.1 GCES01017633 JAR68690.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000192223
+ More
UP000007266 UP000007755 UP000053097 UP000279307 UP000005205 UP000078540 UP000008237 UP000019118 UP000030742 UP000242457 UP000005203 UP000075809 UP000053825 UP000078542 UP000007819 UP000078492 UP000078541 UP000092462 UP000000311 UP000053105 UP000092443 UP000233160 UP000240080 UP000002277 UP000233040 UP000006718 UP000233020 UP000192220 UP000261420 UP000261500 UP000242638 UP000265000
UP000007266 UP000007755 UP000053097 UP000279307 UP000005205 UP000078540 UP000008237 UP000019118 UP000030742 UP000242457 UP000005203 UP000075809 UP000053825 UP000078542 UP000007819 UP000078492 UP000078541 UP000092462 UP000000311 UP000053105 UP000092443 UP000233160 UP000240080 UP000002277 UP000233040 UP000006718 UP000233020 UP000192220 UP000261420 UP000261500 UP000242638 UP000265000
PRIDE
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR036322 WD40_repeat_dom_sf
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR020472 G-protein_beta_WD-40_rep
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR036322 WD40_repeat_dom_sf
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR020472 G-protein_beta_WD-40_rep
Gene 3D
ProteinModelPortal
H9J257
A0A2A4K9C6
A0A0N1INJ2
A0A212EWS3
A0A194PVG0
A0A2H1VC82
+ More
A0A1B6EGH2 A0A1Y1MV48 A0A1W4X8E9 A0A1B6C896 A0A1B6CPU2 A0A1Y1MWA9 D6W864 F4WE20 A0A026WE30 A0A158NUS8 A0A195BBF3 E2BPS6 A0A1W4WXL2 A0A1B6EWN8 A0A1B6EI10 A0A2S2QRQ4 N6TG63 A0A2A3EKG5 A0A088A5V3 A0A151WNS1 A0A0L7R1V1 A0A151IN70 A0A2S2NQX3 J9K1M7 A0A151IVG4 A0A151K056 A0A1B0DNE0 E2A764 A0A0M8ZWE4 A0A131XJE8 A0A1A9Y5P3 A0A2K6FSD5 A0A2K6FSE8 A0A2K6FSD7 A0A2R9C2S8 H2R894 A0A131YVY1 A0A2K5RNK4 F6WVK7 A0A224YXF4 A0A2K5CNV3 L7LX05 A0A1A7W978 A0A1A8CTM6 A0A2I4C2U3 A0A3B4UDC4 A0A3B4UD73 A0A2I4C2T6 A0A3B3VKU7 A0A3P9NPL6 A0A146VH53 A0A146ZQU4
A0A1B6EGH2 A0A1Y1MV48 A0A1W4X8E9 A0A1B6C896 A0A1B6CPU2 A0A1Y1MWA9 D6W864 F4WE20 A0A026WE30 A0A158NUS8 A0A195BBF3 E2BPS6 A0A1W4WXL2 A0A1B6EWN8 A0A1B6EI10 A0A2S2QRQ4 N6TG63 A0A2A3EKG5 A0A088A5V3 A0A151WNS1 A0A0L7R1V1 A0A151IN70 A0A2S2NQX3 J9K1M7 A0A151IVG4 A0A151K056 A0A1B0DNE0 E2A764 A0A0M8ZWE4 A0A131XJE8 A0A1A9Y5P3 A0A2K6FSD5 A0A2K6FSE8 A0A2K6FSD7 A0A2R9C2S8 H2R894 A0A131YVY1 A0A2K5RNK4 F6WVK7 A0A224YXF4 A0A2K5CNV3 L7LX05 A0A1A7W978 A0A1A8CTM6 A0A2I4C2U3 A0A3B4UDC4 A0A3B4UD73 A0A2I4C2T6 A0A3B3VKU7 A0A3P9NPL6 A0A146VH53 A0A146ZQU4
PDB
3ZFD
E-value=9.383e-69,
Score=665
Ontologies
GO
PANTHER
Topology
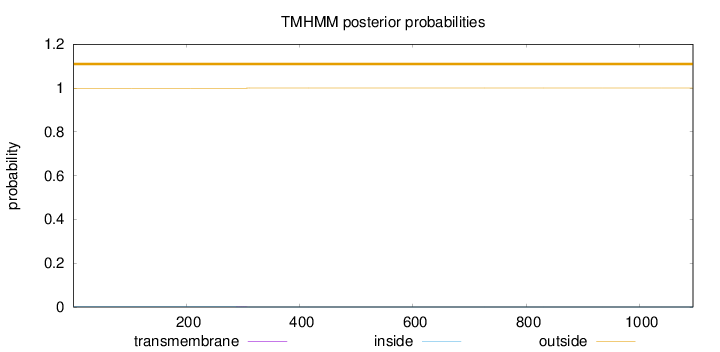
Length:
1094
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01502
Exp number, first 60 AAs:
0.0013
Total prob of N-in:
0.00077
outside
1 - 1094
Population Genetic Test Statistics
Pi
212.365764
Theta
168.379897
Tajima's D
0.957663
CLR
0.377275
CSRT
0.646867656617169
Interpretation
Uncertain
