Gene
KWMTBOMO03013
Pre Gene Modal
BGIBMGA003591
Annotation
Lix1_protein_[Bombyx_mori]
Full name
Protein limb expression 1 homolog
Alternative Name
lowfat
Location in the cell
Cytoplasmic Reliability : 1.475 Mitochondrial Reliability : 1.084
Sequence
CDS
ATGGTGTACCCGGAGCCGTCCCGCTGGGGCGCGCCCGACGCCGCGCTCGCGCCCCTCTACGACGACGTCTACAGAGCTGATGGAGTCCTCTCGACTGTGAATGTAGTGGAAGCCCTCCAAGAGTTTTGGCAGGTGAAGGCGGCACGAGGCGGGGGCGCGGCGGGCGGCGCCCTCGTCATCTACGAGTCTGTGCCGGCTGCTCATCCGCCCTACGTCTGCTACGTCACGCTCCCCGGTGGAGCGTGCTTTGGCAGTTTTCAAAACTGCCCAACTAAAGCGGAAGCAAGACGCAGCGCCGCTAAAATAGCATTAATGAACAGCGTATTCAACGAGCACGAATCTCGGCGGATATCCGACCATTTCATCGAGAAGGCGGTGGCGGAGGCACGCGCGTCCTTCGCCGGAGACACGAGCAACCCACACCAGGACCCCAGCGCTGGTATCGCTGCGTTTAGGTTCATGCTGGAGGCGAACAAAGGTCGGACGATGCTGGAATTCCAGGAGCTGATGACGGTATTCCAGCTGCTCCACTGGAACGGGTCGCTGAGAGCGATGCGCGAGCGGCAATGCTCGCGGCAGGAAGTCGTAGCGCACTACTCGGCCCGGGCACTAGACGACGCGATGCGGGAACAGATGGCGCGAGAATGGGCCACCAGGGAGCGTGAAGCGTCTTCGTCCGGTGGCGGAGTACTACGCGCGGAGCTGGCGAGATCTGAACGCGAGCTTCGGGCCGCGAGGCTGGCGGCACGTGAGCTTCGCTTCCCTAAAGAAAAGCGCGACATCCTCCAACTGGCCGCCCGTCTAGCGCCGCCGCCGCAACCGTGA
Protein
MVYPEPSRWGAPDAALAPLYDDVYRADGVLSTVNVVEALQEFWQVKAARGGGAAGGALVIYESVPAAHPPYVCYVTLPGGACFGSFQNCPTKAEARRSAAKIALMNSVFNEHESRRISDHFIEKAVAEARASFAGDTSNPHQDPSAGIAAFRFMLEANKGRTMLEFQELMTVFQLLHWNGSLRAMRERQCSRQEVVAHYSARALDDAMREQMAREWATREREASSSGGGVLRAELARSERELRAARLAARELRFPKEKRDILQLAARLAPPPQP
Summary
Description
Component of the Fat (ft) signaling pathway that functions in normal development of various organs such as the wing and leg (PubMed:19710173, PubMed:21379328). In developing imaginal disks, involved in regulating both the protein levels and apical localization of ft and ds (PubMed:19710173). Involved in establishing planar cell polarity (PCP) along the anterior-posterior axis of the wing (the early Fz signaling event), probably by acting upstream of ds and ft to regulate Fz activity (PubMed:21379328).
Subunit
Interacts with ft (via intracellular domain) and ds (via intracellular domain).
Similarity
Belongs to the LIX1 family.
Keywords
Cell membrane
Complete proteome
Cytoplasm
Membrane
Reference proteome
Feature
chain Protein limb expression 1 homolog
Uniprot
Q2F5Q3
A0A212EWX0
A0A2A4J3X1
A0A0N1PEU3
A0A2H1VQY9
I4DIQ8
+ More
I4DM31 A0A151JAN9 A0A026WT61 A0A0J7L158 A0A151IEX0 E2AIL0 A0A151I1Y3 A0A0L7LBN1 E9IMI8 A0A151WV28 F4X7U1 A0A195FZ79 E2BM18 A0A1J1IN57 E9G045 A0A3L8DP27 A0A2J7RCF0 A0A0P5EBI1 A0A087ZUU3 A0A154PNP7 A0A1B6D6N0 A0A2A3EQV2 A0A310SQM2 A0A2A3ESF6 A0A0T6AUI0 A0A0N8DPQ9 A0A2P8YNG0 A0A336KUA0 A0A0L7QPJ2 A0A2P2I080 A0A0P5BB77 A0A0P6IA44 A0A067RFD6 Q7PWC8 A0A146LKF0 A0A182QF76 A0A182YFS1 A0A182MGL5 A0A182SUS2 A0A182V565 W5J2B7 A0A182JZY4 A0A182U9X8 A0A182VU23 A0A084VKK4 A0A182X0E1 A0A182LMD9 A0A182FND3 A0A182RLC6 A0A182I7N0 D6W7E9 A0A0P5IR49 E0VQJ9 A0A182PEC8 A0A1B0DPY5 A0A1S4H0M2 A0A1B6IWP6 A0A1B0CHN8 A0A1B6GTP0 A0A182J049 A0A1Y1MXZ2 A0A2M4CX76 A0A336MCM1 A0A0K2UQF6 N6U889 A0A224XVN7 A0A069DRI4 A0A1A9VZR0 B4LVD6 A0A023EYA1 B0WU37 B4JPY5 T1I337 A0A0M4EA86 Q17K63 A0A182G7J9 B4MUC8 A0A1A9Y503 A0A1A9Z9D7 A0A1B0FHP0 A0A0C9R3R7 A0A1B0AWX7 A0A2R5LHD2 A0A0A9VU50 A0A1W4UFI1 B3MPS6 Q9VKY1 B4HWM2 B4Q977 B3N9M7 A0A2R2MN65
I4DM31 A0A151JAN9 A0A026WT61 A0A0J7L158 A0A151IEX0 E2AIL0 A0A151I1Y3 A0A0L7LBN1 E9IMI8 A0A151WV28 F4X7U1 A0A195FZ79 E2BM18 A0A1J1IN57 E9G045 A0A3L8DP27 A0A2J7RCF0 A0A0P5EBI1 A0A087ZUU3 A0A154PNP7 A0A1B6D6N0 A0A2A3EQV2 A0A310SQM2 A0A2A3ESF6 A0A0T6AUI0 A0A0N8DPQ9 A0A2P8YNG0 A0A336KUA0 A0A0L7QPJ2 A0A2P2I080 A0A0P5BB77 A0A0P6IA44 A0A067RFD6 Q7PWC8 A0A146LKF0 A0A182QF76 A0A182YFS1 A0A182MGL5 A0A182SUS2 A0A182V565 W5J2B7 A0A182JZY4 A0A182U9X8 A0A182VU23 A0A084VKK4 A0A182X0E1 A0A182LMD9 A0A182FND3 A0A182RLC6 A0A182I7N0 D6W7E9 A0A0P5IR49 E0VQJ9 A0A182PEC8 A0A1B0DPY5 A0A1S4H0M2 A0A1B6IWP6 A0A1B0CHN8 A0A1B6GTP0 A0A182J049 A0A1Y1MXZ2 A0A2M4CX76 A0A336MCM1 A0A0K2UQF6 N6U889 A0A224XVN7 A0A069DRI4 A0A1A9VZR0 B4LVD6 A0A023EYA1 B0WU37 B4JPY5 T1I337 A0A0M4EA86 Q17K63 A0A182G7J9 B4MUC8 A0A1A9Y503 A0A1A9Z9D7 A0A1B0FHP0 A0A0C9R3R7 A0A1B0AWX7 A0A2R5LHD2 A0A0A9VU50 A0A1W4UFI1 B3MPS6 Q9VKY1 B4HWM2 B4Q977 B3N9M7 A0A2R2MN65
Pubmed
22118469
26354079
22651552
24508170
20798317
26227816
+ More
21282665 21719571 21292972 30249741 29403074 24845553 12364791 26823975 25244985 20920257 23761445 24438588 20966253 18362917 19820115 20566863 28004739 23537049 26334808 17994087 18057021 25474469 17510324 26483478 25401762 10731132 12537572 19710173 21379328 22936249
21282665 21719571 21292972 30249741 29403074 24845553 12364791 26823975 25244985 20920257 23761445 24438588 20966253 18362917 19820115 20566863 28004739 23537049 26334808 17994087 18057021 25474469 17510324 26483478 25401762 10731132 12537572 19710173 21379328 22936249
EMBL
DQ311370
ABD36314.1
AGBW02011913
OWR45934.1
NWSH01003158
PCG66847.1
+ More
KQ461187 KPJ07355.1 ODYU01003888 SOQ43208.1 AK401176 KQ459597 BAM17798.1 KPI95113.1 AK402349 BAM18971.1 KQ979254 KYN22101.1 KK107128 EZA58294.1 LBMM01001492 KMQ96214.1 KQ977835 KYM99367.1 GL439822 EFN66731.1 KQ976554 KYM80818.1 JTDY01001815 KOB72804.1 GL764165 EFZ18213.1 KQ982709 KYQ51779.1 GL888900 EGI57427.1 KQ981193 KYN45109.1 GL449100 EFN83267.1 CVRI01000055 CRL01186.1 GL732528 EFX86835.1 QOIP01000006 RLU21629.1 NEVH01005885 PNF38500.1 GDIP01143760 LRGB01001374 JAJ79642.1 KZS12115.1 KQ435007 KZC13505.1 GEDC01015968 JAS21330.1 KZ288193 PBC34145.1 KQ760995 OAD58510.1 PBC34146.1 LJIG01022781 KRT78748.1 GDIP01119261 GDIP01012975 JAM90740.1 PYGN01000469 PSN45782.1 UFQS01000934 UFQT01000934 SSX07842.1 SSX28076.1 KQ414832 KOC60396.1 IACF01001723 LAB67409.1 GDIP01187400 JAJ36002.1 GDIQ01007448 JAN87289.1 KK852506 KDR22477.1 AAAB01008984 EAA14824.3 GDHC01010118 JAQ08511.1 AXCN02000625 AXCM01000226 ADMH02002194 ETN57881.1 ATLV01014228 KE524948 KFB38498.1 APCN01003430 KQ971307 EFA11217.2 GDIQ01210214 JAK41511.1 DS235430 EEB15655.1 AJVK01008339 GECU01016345 JAS91361.1 AJWK01012546 GECZ01003988 JAS65781.1 GEZM01020526 JAV89255.1 GGFL01005300 MBW69478.1 SSX07840.1 SSX28074.1 HACA01023112 CDW40473.1 APGK01039147 KB740967 KB632399 ENN76866.1 ERL94646.1 GFTR01004253 JAW12173.1 GBGD01002588 JAC86301.1 CH940649 EDW63315.1 KRF81035.1 KRF81036.1 GBBI01004655 JAC14057.1 DS232100 EDS34756.1 CH916372 EDV98965.1 ACPB03024256 CP012523 ALC39264.1 CH477228 EAT47029.1 JXUM01046495 JXUM01046496 JXUM01079389 KQ563124 KQ561479 KXJ74436.1 KXJ78451.1 CH963857 EDW76054.1 CCAG010020380 GBYB01010968 GBYB01010969 JAG80735.1 JAG80736.1 JXJN01004959 GGLE01004808 MBY08934.1 GBHO01043797 GBHO01027905 GBRD01009634 GDHC01002236 JAF99806.1 JAG15699.1 JAG56190.1 JAQ16393.1 CH902620 EDV32324.1 KPU73900.1 AE014134 BT024350 ABC86412.1 CH480818 EDW52417.1 CM000361 CM002910 EDX04519.1 KMY89503.1 CH954177 EDV58522.1 KQS70432.1
KQ461187 KPJ07355.1 ODYU01003888 SOQ43208.1 AK401176 KQ459597 BAM17798.1 KPI95113.1 AK402349 BAM18971.1 KQ979254 KYN22101.1 KK107128 EZA58294.1 LBMM01001492 KMQ96214.1 KQ977835 KYM99367.1 GL439822 EFN66731.1 KQ976554 KYM80818.1 JTDY01001815 KOB72804.1 GL764165 EFZ18213.1 KQ982709 KYQ51779.1 GL888900 EGI57427.1 KQ981193 KYN45109.1 GL449100 EFN83267.1 CVRI01000055 CRL01186.1 GL732528 EFX86835.1 QOIP01000006 RLU21629.1 NEVH01005885 PNF38500.1 GDIP01143760 LRGB01001374 JAJ79642.1 KZS12115.1 KQ435007 KZC13505.1 GEDC01015968 JAS21330.1 KZ288193 PBC34145.1 KQ760995 OAD58510.1 PBC34146.1 LJIG01022781 KRT78748.1 GDIP01119261 GDIP01012975 JAM90740.1 PYGN01000469 PSN45782.1 UFQS01000934 UFQT01000934 SSX07842.1 SSX28076.1 KQ414832 KOC60396.1 IACF01001723 LAB67409.1 GDIP01187400 JAJ36002.1 GDIQ01007448 JAN87289.1 KK852506 KDR22477.1 AAAB01008984 EAA14824.3 GDHC01010118 JAQ08511.1 AXCN02000625 AXCM01000226 ADMH02002194 ETN57881.1 ATLV01014228 KE524948 KFB38498.1 APCN01003430 KQ971307 EFA11217.2 GDIQ01210214 JAK41511.1 DS235430 EEB15655.1 AJVK01008339 GECU01016345 JAS91361.1 AJWK01012546 GECZ01003988 JAS65781.1 GEZM01020526 JAV89255.1 GGFL01005300 MBW69478.1 SSX07840.1 SSX28074.1 HACA01023112 CDW40473.1 APGK01039147 KB740967 KB632399 ENN76866.1 ERL94646.1 GFTR01004253 JAW12173.1 GBGD01002588 JAC86301.1 CH940649 EDW63315.1 KRF81035.1 KRF81036.1 GBBI01004655 JAC14057.1 DS232100 EDS34756.1 CH916372 EDV98965.1 ACPB03024256 CP012523 ALC39264.1 CH477228 EAT47029.1 JXUM01046495 JXUM01046496 JXUM01079389 KQ563124 KQ561479 KXJ74436.1 KXJ78451.1 CH963857 EDW76054.1 CCAG010020380 GBYB01010968 GBYB01010969 JAG80735.1 JAG80736.1 JXJN01004959 GGLE01004808 MBY08934.1 GBHO01043797 GBHO01027905 GBRD01009634 GDHC01002236 JAF99806.1 JAG15699.1 JAG56190.1 JAQ16393.1 CH902620 EDV32324.1 KPU73900.1 AE014134 BT024350 ABC86412.1 CH480818 EDW52417.1 CM000361 CM002910 EDX04519.1 KMY89503.1 CH954177 EDV58522.1 KQS70432.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000053268
UP000078492
UP000053097
+ More
UP000036403 UP000078542 UP000000311 UP000078540 UP000037510 UP000075809 UP000007755 UP000078541 UP000008237 UP000183832 UP000000305 UP000279307 UP000235965 UP000076858 UP000005203 UP000076502 UP000242457 UP000245037 UP000053825 UP000027135 UP000007062 UP000075886 UP000076408 UP000075883 UP000075901 UP000075903 UP000000673 UP000075881 UP000075902 UP000075920 UP000030765 UP000076407 UP000075882 UP000069272 UP000075900 UP000075840 UP000007266 UP000009046 UP000075885 UP000092462 UP000092461 UP000075880 UP000019118 UP000030742 UP000091820 UP000008792 UP000002320 UP000001070 UP000015103 UP000092553 UP000008820 UP000069940 UP000249989 UP000007798 UP000092443 UP000092445 UP000092444 UP000092460 UP000192221 UP000007801 UP000000803 UP000001292 UP000000304 UP000008711 UP000085678
UP000036403 UP000078542 UP000000311 UP000078540 UP000037510 UP000075809 UP000007755 UP000078541 UP000008237 UP000183832 UP000000305 UP000279307 UP000235965 UP000076858 UP000005203 UP000076502 UP000242457 UP000245037 UP000053825 UP000027135 UP000007062 UP000075886 UP000076408 UP000075883 UP000075901 UP000075903 UP000000673 UP000075881 UP000075902 UP000075920 UP000030765 UP000076407 UP000075882 UP000069272 UP000075900 UP000075840 UP000007266 UP000009046 UP000075885 UP000092462 UP000092461 UP000075880 UP000019118 UP000030742 UP000091820 UP000008792 UP000002320 UP000001070 UP000015103 UP000092553 UP000008820 UP000069940 UP000249989 UP000007798 UP000092443 UP000092445 UP000092444 UP000092460 UP000192221 UP000007801 UP000000803 UP000001292 UP000000304 UP000008711 UP000085678
Pfam
PF14954 LIX1
Interpro
IPR029270
LIX1
ProteinModelPortal
Q2F5Q3
A0A212EWX0
A0A2A4J3X1
A0A0N1PEU3
A0A2H1VQY9
I4DIQ8
+ More
I4DM31 A0A151JAN9 A0A026WT61 A0A0J7L158 A0A151IEX0 E2AIL0 A0A151I1Y3 A0A0L7LBN1 E9IMI8 A0A151WV28 F4X7U1 A0A195FZ79 E2BM18 A0A1J1IN57 E9G045 A0A3L8DP27 A0A2J7RCF0 A0A0P5EBI1 A0A087ZUU3 A0A154PNP7 A0A1B6D6N0 A0A2A3EQV2 A0A310SQM2 A0A2A3ESF6 A0A0T6AUI0 A0A0N8DPQ9 A0A2P8YNG0 A0A336KUA0 A0A0L7QPJ2 A0A2P2I080 A0A0P5BB77 A0A0P6IA44 A0A067RFD6 Q7PWC8 A0A146LKF0 A0A182QF76 A0A182YFS1 A0A182MGL5 A0A182SUS2 A0A182V565 W5J2B7 A0A182JZY4 A0A182U9X8 A0A182VU23 A0A084VKK4 A0A182X0E1 A0A182LMD9 A0A182FND3 A0A182RLC6 A0A182I7N0 D6W7E9 A0A0P5IR49 E0VQJ9 A0A182PEC8 A0A1B0DPY5 A0A1S4H0M2 A0A1B6IWP6 A0A1B0CHN8 A0A1B6GTP0 A0A182J049 A0A1Y1MXZ2 A0A2M4CX76 A0A336MCM1 A0A0K2UQF6 N6U889 A0A224XVN7 A0A069DRI4 A0A1A9VZR0 B4LVD6 A0A023EYA1 B0WU37 B4JPY5 T1I337 A0A0M4EA86 Q17K63 A0A182G7J9 B4MUC8 A0A1A9Y503 A0A1A9Z9D7 A0A1B0FHP0 A0A0C9R3R7 A0A1B0AWX7 A0A2R5LHD2 A0A0A9VU50 A0A1W4UFI1 B3MPS6 Q9VKY1 B4HWM2 B4Q977 B3N9M7 A0A2R2MN65
I4DM31 A0A151JAN9 A0A026WT61 A0A0J7L158 A0A151IEX0 E2AIL0 A0A151I1Y3 A0A0L7LBN1 E9IMI8 A0A151WV28 F4X7U1 A0A195FZ79 E2BM18 A0A1J1IN57 E9G045 A0A3L8DP27 A0A2J7RCF0 A0A0P5EBI1 A0A087ZUU3 A0A154PNP7 A0A1B6D6N0 A0A2A3EQV2 A0A310SQM2 A0A2A3ESF6 A0A0T6AUI0 A0A0N8DPQ9 A0A2P8YNG0 A0A336KUA0 A0A0L7QPJ2 A0A2P2I080 A0A0P5BB77 A0A0P6IA44 A0A067RFD6 Q7PWC8 A0A146LKF0 A0A182QF76 A0A182YFS1 A0A182MGL5 A0A182SUS2 A0A182V565 W5J2B7 A0A182JZY4 A0A182U9X8 A0A182VU23 A0A084VKK4 A0A182X0E1 A0A182LMD9 A0A182FND3 A0A182RLC6 A0A182I7N0 D6W7E9 A0A0P5IR49 E0VQJ9 A0A182PEC8 A0A1B0DPY5 A0A1S4H0M2 A0A1B6IWP6 A0A1B0CHN8 A0A1B6GTP0 A0A182J049 A0A1Y1MXZ2 A0A2M4CX76 A0A336MCM1 A0A0K2UQF6 N6U889 A0A224XVN7 A0A069DRI4 A0A1A9VZR0 B4LVD6 A0A023EYA1 B0WU37 B4JPY5 T1I337 A0A0M4EA86 Q17K63 A0A182G7J9 B4MUC8 A0A1A9Y503 A0A1A9Z9D7 A0A1B0FHP0 A0A0C9R3R7 A0A1B0AWX7 A0A2R5LHD2 A0A0A9VU50 A0A1W4UFI1 B3MPS6 Q9VKY1 B4HWM2 B4Q977 B3N9M7 A0A2R2MN65
Ontologies
Topology
Subcellular location
Apical cell membrane
In wing imaginal disks, strong cytoplasmic expression in the wing pouch but in regions of the wing hinge preferentially expressed in the sub-apical membrane. With evidence from 1 publications.
Cytoplasm In wing imaginal disks, strong cytoplasmic expression in the wing pouch but in regions of the wing hinge preferentially expressed in the sub-apical membrane. With evidence from 1 publications.
Cytoplasm In wing imaginal disks, strong cytoplasmic expression in the wing pouch but in regions of the wing hinge preferentially expressed in the sub-apical membrane. With evidence from 1 publications.
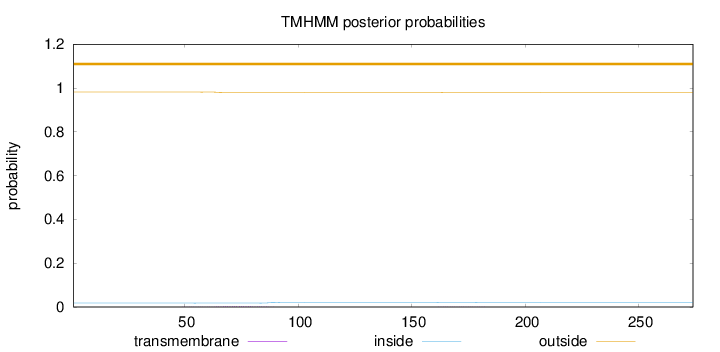
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07636
Exp number, first 60 AAs:
0.00601
Total prob of N-in:
0.01830
outside
1 - 274
Population Genetic Test Statistics
Pi
22.019904
Theta
185.682917
Tajima's D
0.052781
CLR
1448.31441
CSRT
0.381330933453327
Interpretation
Uncertain
