Gene
KWMTBOMO03011
Pre Gene Modal
BGIBMGA003589
Annotation
PREDICTED:_uncharacterized_protein_LOC106099267_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 2.538
Sequence
CDS
ATGCCTATTCGAATACAAATATTTCAAATAGGTTACTACTCCGTCAAAACTATTATACCTACTAGGGAGGCGCATAGGCTGGGCCGGTCTGCGGCGAGCGAGCGTATGGCTCTGCTGTACCGAGCGACGCGGCTGAAGGACCGCGCCGACACCCATGTCTTCACTTTTGTGGTCACGCGCTCCGCCACCAGGGAGCCCGACCGCGATGTCACCTCCAAGGACTTCTGCTGCGCCCACCAGAGATGGGCGGTCGCCTTCAGCCGCACTGACACCTCACTCGGTGTATATCTGGTGTGGAGAGGAGCGTGCGAAGGAATGAGGGTTTACGTAGACTTCACTTTTACTCTTCTAAGCCGTGACCATTTTACAGCCAACGAAGGCTTCTCCGGCAAGCAGGTTCGGTTCAGCGCCGGATGCGCTGCCCAAGGTCGCGGCAGATGCGTGTCGCTCGCGGAGCTTAACGCCAAGTTCGCGGACGCACGCGGCGAATTTCAACTTGAACTTACTATGTCCCGAGTGCGCACTCTCTACTCTTGCGAGATACGCGCGCCTCGCCTTGATACTCCGCCAATAGCATTTGCCGGCTTCGACTGGCAAGTTTCAGTCAGCGGCGGGAACAAAGAACCCTTAGCTTTACGTCTCGTTCGATTGTCTGGCGAAGGGCAGAAATGCCGAGTTCGTTATGCCCTTGCGCTTGGCGAGGGCGAGCGGCGTATACACTCAGGACCTCTAGAGTGTGTCTGTGATTCTGAGGGACGGACCCCGCCTTGGAACCCTCGTCCGCCTTCTCGCCTACTGCACAAAGGCGTCCGACTTACTGTAGAACTGGTTTGGGCGCGAGCAGTCGCAGAGTTGGCGGTTTCGGCTTCGGGAAGAGCAGCCACTTGCTATGATCGTGATAAGCAAGCGTGGGCACTTCGGTGCGACATGCATTCGGAAACAGTGCGTCTGCACATGCTGTATAGAGATGTGCATCACGTCCCGCGCAACCACCTGCGTTACGTCAGCTGGTCGGCTTGGTTGGTTCGCGCTACTCCTGCAGACTCGGAATCTGAGGCAGAGGAACTTCCTGGAGCTCCTTTCGAACATTACTACGCTCAAGACAGTGCTGATGAGGGTCTCATGATGGAGACCGCTTTGCGAGTGGAAGACGTTTCACGAGCCGGTTGTTCTTTTATGCACCCTGGAGGGGAACTTCGCGTACGCCTCGAATGGGGCGACACCTATCTACTCTTTCAAGCAACTTATCACGTTTATGATGATTTGTGCCGTTTACATGCCCACCAAATGAGACGGGAGATTGCAGCACTTCAGGCGGAAAATTATTCATTGGAGCGACAGTTGTTTAGCTATCAGAAGAGTCTGGCCTATGCTCAAGCACAGGCGGAGGAGCCACCGGTACCGGAACCCGGCGGACGGCGGTCGCCGGCTGAACGCTCCCTCTCTACAGACACGGAGTACGCGTGA
Protein
MPIRIQIFQIGYYSVKTIIPTREAHRLGRSAASERMALLYRATRLKDRADTHVFTFVVTRSATREPDRDVTSKDFCCAHQRWAVAFSRTDTSLGVYLVWRGACEGMRVYVDFTFTLLSRDHFTANEGFSGKQVRFSAGCAAQGRGRCVSLAELNAKFADARGEFQLELTMSRVRTLYSCEIRAPRLDTPPIAFAGFDWQVSVSGGNKEPLALRLVRLSGEGQKCRVRYALALGEGERRIHSGPLECVCDSEGRTPPWNPRPPSRLLHKGVRLTVELVWARAVAELAVSASGRAATCYDRDKQAWALRCDMHSETVRLHMLYRDVHHVPRNHLRYVSWSAWLVRATPADSESEAEELPGAPFEHYYAQDSADEGLMMETALRVEDVSRAGCSFMHPGGELRVRLEWGDTYLLFQATYHVYDDLCRLHAHQMRREIAALQAENYSLERQLFSYQKSLAYAQAQAEEPPVPEPGGRRSPAERSLSTDTEYA
Summary
Uniprot
H9J252
A0A194PNV5
A0A0N0PAM1
A0A2A4J446
A0A2H1V2X7
A0A212EWR6
+ More
A0A0L7LP43 A0A0C9QE22 A0A154P365 E2AQ15 A0A3L8DIN1 A0A026WMF0 A0A158NKX9 A0A151I276 A0A088A0B5 F4WAC3 A0A195CSG0 A0A151WVJ4 A0A195E3G9 A0A195FRT5 K7JU74 A0A232ERV7 E2BEL6 E9IZ15 T1HT81 A0A2A3ERY4 A0A0M8ZQA7 A0A2S2PCS9 A0A2S2QJW0 A0A2J7PZ44 A0A2S2NR05 A0A2S2Q4S6 A0A226F5K0 A0A147BG41 T1IZ86 A0A2P8ZC67 A0A0J7L5R5 A0A1D2M6C8 A0A087UWB6 A0A132AFX6 A0A2P6KLA3 A0A2T7NIW7 A0A2C9K432 B7PSE9 A0A132AHU2 V3ZUH9 A0A210PWD0 E0VRT9 A0A1S3K6U9 A0A1V9X766 A0A0L8FYW7 A0A3M7RC09 A0A1W0WZS0 A0A1D1V9U2 C3ZBW8 A0A0V0YA43
A0A0L7LP43 A0A0C9QE22 A0A154P365 E2AQ15 A0A3L8DIN1 A0A026WMF0 A0A158NKX9 A0A151I276 A0A088A0B5 F4WAC3 A0A195CSG0 A0A151WVJ4 A0A195E3G9 A0A195FRT5 K7JU74 A0A232ERV7 E2BEL6 E9IZ15 T1HT81 A0A2A3ERY4 A0A0M8ZQA7 A0A2S2PCS9 A0A2S2QJW0 A0A2J7PZ44 A0A2S2NR05 A0A2S2Q4S6 A0A226F5K0 A0A147BG41 T1IZ86 A0A2P8ZC67 A0A0J7L5R5 A0A1D2M6C8 A0A087UWB6 A0A132AFX6 A0A2P6KLA3 A0A2T7NIW7 A0A2C9K432 B7PSE9 A0A132AHU2 V3ZUH9 A0A210PWD0 E0VRT9 A0A1S3K6U9 A0A1V9X766 A0A0L8FYW7 A0A3M7RC09 A0A1W0WZS0 A0A1D1V9U2 C3ZBW8 A0A0V0YA43
Pubmed
EMBL
BABH01007413
KQ459597
KPI95111.1
KQ461187
KPJ07353.1
NWSH01003158
+ More
PCG66845.1 ODYU01000223 SOQ34634.1 AGBW02011913 OWR45936.1 JTDY01000434 KOB77190.1 GBYB01001564 JAG71331.1 KQ434809 KZC06272.1 GL441649 EFN64472.1 QOIP01000007 RLU20062.1 KK107152 EZA57200.1 ADTU01019003 KQ976551 KYM80851.1 GL888048 EGI68806.1 KQ977306 KYN03648.1 KQ982718 KYQ51695.1 KQ979701 KYN19683.1 KQ981281 KYN43315.1 AAZX01006212 NNAY01002549 OXU21077.1 GL447829 EFN85850.1 GL767078 EFZ14166.1 ACPB03002586 KZ288194 PBC33976.1 KQ436237 KOX67389.1 GGMR01014127 MBY26746.1 GGMS01008846 MBY78049.1 NEVH01020340 PNF21605.1 GGMR01006938 MBY19557.1 GGMS01003526 MBY72729.1 LNIX01000001 OXA65069.1 GEGO01005651 JAR89753.1 AFFK01020413 PYGN01000108 PSN54089.1 LBMM01000623 KMQ97956.1 LJIJ01003451 ODM88537.1 KK121986 KFM81655.1 JXLN01014224 KPM09906.1 MWRG01009369 PRD27125.1 PZQS01000012 PVD21119.1 ABJB010301353 ABJB010301523 ABJB010349937 DS778349 EEC09521.1 JXLN01014390 KPM10010.1 KB201701 ESO95143.1 NEDP02005445 OWF40798.1 DS235562 EEB16095.1 MNPL01022065 OQR69122.1 KQ425203 KOF69774.1 REGN01003743 RNA21047.1 MTYJ01000028 OQV20710.1 BDGG01000003 GAU95623.1 GG666603 EEN50355.1 JYDU01000036 KRX97034.1
PCG66845.1 ODYU01000223 SOQ34634.1 AGBW02011913 OWR45936.1 JTDY01000434 KOB77190.1 GBYB01001564 JAG71331.1 KQ434809 KZC06272.1 GL441649 EFN64472.1 QOIP01000007 RLU20062.1 KK107152 EZA57200.1 ADTU01019003 KQ976551 KYM80851.1 GL888048 EGI68806.1 KQ977306 KYN03648.1 KQ982718 KYQ51695.1 KQ979701 KYN19683.1 KQ981281 KYN43315.1 AAZX01006212 NNAY01002549 OXU21077.1 GL447829 EFN85850.1 GL767078 EFZ14166.1 ACPB03002586 KZ288194 PBC33976.1 KQ436237 KOX67389.1 GGMR01014127 MBY26746.1 GGMS01008846 MBY78049.1 NEVH01020340 PNF21605.1 GGMR01006938 MBY19557.1 GGMS01003526 MBY72729.1 LNIX01000001 OXA65069.1 GEGO01005651 JAR89753.1 AFFK01020413 PYGN01000108 PSN54089.1 LBMM01000623 KMQ97956.1 LJIJ01003451 ODM88537.1 KK121986 KFM81655.1 JXLN01014224 KPM09906.1 MWRG01009369 PRD27125.1 PZQS01000012 PVD21119.1 ABJB010301353 ABJB010301523 ABJB010349937 DS778349 EEC09521.1 JXLN01014390 KPM10010.1 KB201701 ESO95143.1 NEDP02005445 OWF40798.1 DS235562 EEB16095.1 MNPL01022065 OQR69122.1 KQ425203 KOF69774.1 REGN01003743 RNA21047.1 MTYJ01000028 OQV20710.1 BDGG01000003 GAU95623.1 GG666603 EEN50355.1 JYDU01000036 KRX97034.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000076502 UP000000311 UP000279307 UP000053097 UP000005205 UP000078540 UP000005203 UP000007755 UP000078542 UP000075809 UP000078492 UP000078541 UP000002358 UP000215335 UP000008237 UP000015103 UP000242457 UP000053105 UP000235965 UP000198287 UP000245037 UP000036403 UP000094527 UP000054359 UP000245119 UP000076420 UP000001555 UP000030746 UP000242188 UP000009046 UP000085678 UP000192247 UP000053454 UP000276133 UP000186922 UP000001554 UP000054815
UP000076502 UP000000311 UP000279307 UP000053097 UP000005205 UP000078540 UP000005203 UP000007755 UP000078542 UP000075809 UP000078492 UP000078541 UP000002358 UP000215335 UP000008237 UP000015103 UP000242457 UP000053105 UP000235965 UP000198287 UP000245037 UP000036403 UP000094527 UP000054359 UP000245119 UP000076420 UP000001555 UP000030746 UP000242188 UP000009046 UP000085678 UP000192247 UP000053454 UP000276133 UP000186922 UP000001554 UP000054815
PRIDE
Pfam
PF00917 MATH
SUPFAM
SSF49599
SSF49599
Gene 3D
CDD
ProteinModelPortal
H9J252
A0A194PNV5
A0A0N0PAM1
A0A2A4J446
A0A2H1V2X7
A0A212EWR6
+ More
A0A0L7LP43 A0A0C9QE22 A0A154P365 E2AQ15 A0A3L8DIN1 A0A026WMF0 A0A158NKX9 A0A151I276 A0A088A0B5 F4WAC3 A0A195CSG0 A0A151WVJ4 A0A195E3G9 A0A195FRT5 K7JU74 A0A232ERV7 E2BEL6 E9IZ15 T1HT81 A0A2A3ERY4 A0A0M8ZQA7 A0A2S2PCS9 A0A2S2QJW0 A0A2J7PZ44 A0A2S2NR05 A0A2S2Q4S6 A0A226F5K0 A0A147BG41 T1IZ86 A0A2P8ZC67 A0A0J7L5R5 A0A1D2M6C8 A0A087UWB6 A0A132AFX6 A0A2P6KLA3 A0A2T7NIW7 A0A2C9K432 B7PSE9 A0A132AHU2 V3ZUH9 A0A210PWD0 E0VRT9 A0A1S3K6U9 A0A1V9X766 A0A0L8FYW7 A0A3M7RC09 A0A1W0WZS0 A0A1D1V9U2 C3ZBW8 A0A0V0YA43
A0A0L7LP43 A0A0C9QE22 A0A154P365 E2AQ15 A0A3L8DIN1 A0A026WMF0 A0A158NKX9 A0A151I276 A0A088A0B5 F4WAC3 A0A195CSG0 A0A151WVJ4 A0A195E3G9 A0A195FRT5 K7JU74 A0A232ERV7 E2BEL6 E9IZ15 T1HT81 A0A2A3ERY4 A0A0M8ZQA7 A0A2S2PCS9 A0A2S2QJW0 A0A2J7PZ44 A0A2S2NR05 A0A2S2Q4S6 A0A226F5K0 A0A147BG41 T1IZ86 A0A2P8ZC67 A0A0J7L5R5 A0A1D2M6C8 A0A087UWB6 A0A132AFX6 A0A2P6KLA3 A0A2T7NIW7 A0A2C9K432 B7PSE9 A0A132AHU2 V3ZUH9 A0A210PWD0 E0VRT9 A0A1S3K6U9 A0A1V9X766 A0A0L8FYW7 A0A3M7RC09 A0A1W0WZS0 A0A1D1V9U2 C3ZBW8 A0A0V0YA43
Ontologies
GO
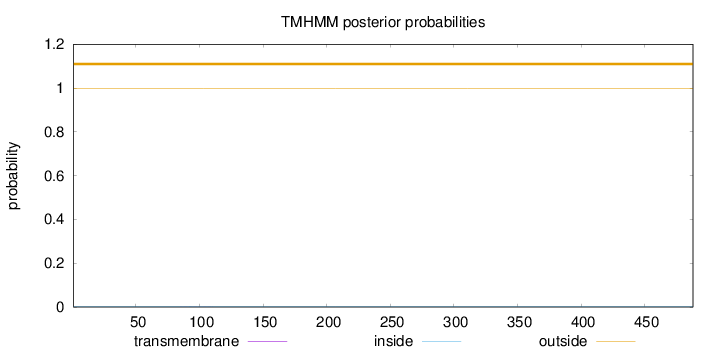
Topology
Length:
488
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00818999999999998
Exp number, first 60 AAs:
0.00089
Total prob of N-in:
0.00218
outside
1 - 488
Population Genetic Test Statistics
Pi
177.145858
Theta
177.868133
Tajima's D
-0.386146
CLR
1.142762
CSRT
0.269736513174341
Interpretation
Uncertain
