Gene
KWMTBOMO03008
Pre Gene Modal
BGIBMGA003606
Annotation
PREDICTED:_nucleoside_diphosphate-linked_moiety_X_motif_19?_mitochondrial-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.522 Nuclear Reliability : 1.152
Sequence
CDS
ATGGCAACACTCGGTAAACGTACTCGGACGGGTCGCGGTGCGTTTCCAAACAGCGTGGTGTTCCCCGGCGGAGTTACCGAAGACGCTGACGAAGACGATCGCTGGCTACAGCTGTTCAGTTTGTTTGGTTACACGAACACAGATTTAGAGTCTCTACATCACGCTAATGGTCCTATCACTCCAATTTTACAAAAGAATCCCATTCGAAGGCACATATCATTAAGAATTACAGCCATCAGGGAAACTTTTGAAGAGTTAGGGATTCTGCTATGCAGTCAGCAGCACAAGAAGCAAAAGGACGGCTTAAGGGCGGATTTCATAGCTAATATTGATGTTAAAACTTGGCAGGACAGAGTAAGCAAGAATCCAGAAAGCCTTTGGAACCTGTGTGAAGATTATAAGTGCTATCCAGACATATGGTCACTACATTACTGGAGCAATTGGTTAACTCCAGCCACTTTACCGAAGAGATTCAACACAGCCTTCTTTGTGACAGCGTTAGAGGAAAAGCCTGAGTTAAAAAATTACTCTACAGAAGTTGCCTTTGTTAAGTGGTCAGACCCAACTGAGATACTCAACAGCAGTGACGTGAAAGCAACAGTTGTTATGGTCACGAACTTGTCTTTCCTGTGTACGTAA
Protein
MATLGKRTRTGRGAFPNSVVFPGGVTEDADEDDRWLQLFSLFGYTNTDLESLHHANGPITPILQKNPIRRHISLRITAIRETFEELGILLCSQQHKKQKDGLRADFIANIDVKTWQDRVSKNPESLWNLCEDYKCYPDIWSLHYWSNWLTPATLPKRFNTAFFVTALEEKPELKNYSTEVAFVKWSDPTEILNSSDVKATVVMVTNLSFLCT
Summary
Uniprot
H9J269
I4DPG6
A0A194PVF5
A0A0N1I4S7
A0A2A4J515
A0A212FJW5
+ More
A0A1B0FNM7 D3TL26 A0A2P8YMX7 A0A1B0A518 A0A1B0BJG1 A0A2J7PZQ0 A0A0C9R086 A0A1A9V9K8 A0A026WP48 A0A232F570 A0A0L7R6Q5 A0A1Y1L567 A0A0T6BB49 B4I5W1 A0A154P563 A0A0T6AZS1 E2B2T2 A0A088AGP6 A0A1Y1L0L0 Q9VIV6 A0A1Y1L0E2 A0A1I8MI79 E9ICM1 B4Q9Y8 B4PAZ4 B3MN59 T1PJ57 A0A2A3E9X4 A0A0P8XFP8 A0A3B0KBE1 B3NM47 A0A1L8DAT2 Q29K58 B4GWX8 A0A1I8PRM5 B4M8J6 A0A1L8DAD0 A0A0M4E5Q9 A0A1I8PRR9 A0A1W4VG39 A0A195CUC9 A0A158NQ78 A0A1Y1L0D4 B4JCV6 A0A0J7NHD2 E2A5K3 A0A1I8M2J5 A0A1I8NUH6 A0A2B4RLM4 A0A195DER3 W8C7D1 A0A3M6UHR7 A0A1I8Q5P3 F4X5I6 A0A1B0D1D1 A0A034WD54 B4MYR4 A0A1I8MFU3 A0A0P5C3X5 A0A0N7ZUY7 A0A0P6EDN2 A0A0P5LD38 A0A0P6DDT0 K7IV53 A0A0P4Y577 A0A0P5C827 A0A0P5GS02 A0A0P5KFN9 A0A0P5DRG9 A0A0P5H4F7 A0A195FQE5 A0A0P6H1A2 B0W4P1 A0A0P5YTT5 A0A162SSA6 A0A0P6CRI3 G3PVB0 A0A0L0BQ83 A0A195BJ86 A0A1Q3FJ93 A0A151WYH1 E0VQL8 A0A1I8P8E6 A0A182YMQ6 C3ZBL8 A0A0P4YTT0 C3ZIX2 B3MN62 A0A0A1XLV1 A0A2I4CKG6 I3J2E7 A0A0P5J495
A0A1B0FNM7 D3TL26 A0A2P8YMX7 A0A1B0A518 A0A1B0BJG1 A0A2J7PZQ0 A0A0C9R086 A0A1A9V9K8 A0A026WP48 A0A232F570 A0A0L7R6Q5 A0A1Y1L567 A0A0T6BB49 B4I5W1 A0A154P563 A0A0T6AZS1 E2B2T2 A0A088AGP6 A0A1Y1L0L0 Q9VIV6 A0A1Y1L0E2 A0A1I8MI79 E9ICM1 B4Q9Y8 B4PAZ4 B3MN59 T1PJ57 A0A2A3E9X4 A0A0P8XFP8 A0A3B0KBE1 B3NM47 A0A1L8DAT2 Q29K58 B4GWX8 A0A1I8PRM5 B4M8J6 A0A1L8DAD0 A0A0M4E5Q9 A0A1I8PRR9 A0A1W4VG39 A0A195CUC9 A0A158NQ78 A0A1Y1L0D4 B4JCV6 A0A0J7NHD2 E2A5K3 A0A1I8M2J5 A0A1I8NUH6 A0A2B4RLM4 A0A195DER3 W8C7D1 A0A3M6UHR7 A0A1I8Q5P3 F4X5I6 A0A1B0D1D1 A0A034WD54 B4MYR4 A0A1I8MFU3 A0A0P5C3X5 A0A0N7ZUY7 A0A0P6EDN2 A0A0P5LD38 A0A0P6DDT0 K7IV53 A0A0P4Y577 A0A0P5C827 A0A0P5GS02 A0A0P5KFN9 A0A0P5DRG9 A0A0P5H4F7 A0A195FQE5 A0A0P6H1A2 B0W4P1 A0A0P5YTT5 A0A162SSA6 A0A0P6CRI3 G3PVB0 A0A0L0BQ83 A0A195BJ86 A0A1Q3FJ93 A0A151WYH1 E0VQL8 A0A1I8P8E6 A0A182YMQ6 C3ZBL8 A0A0P4YTT0 C3ZIX2 B3MN62 A0A0A1XLV1 A0A2I4CKG6 I3J2E7 A0A0P5J495
Pubmed
19121390
22651552
26354079
22118469
20353571
29403074
+ More
24508170 30249741 28648823 28004739 17994087 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 21282665 22936249 17550304 15632085 21347285 24495485 30382153 21719571 25348373 20075255 26108605 20566863 25244985 18563158 25830018 25186727
24508170 30249741 28648823 28004739 17994087 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 21282665 22936249 17550304 15632085 21347285 24495485 30382153 21719571 25348373 20075255 26108605 20566863 25244985 18563158 25830018 25186727
EMBL
BABH01007424
AK403590
BAM19806.1
KQ459597
KPI95110.1
KQ461187
+ More
KPJ07352.1 NWSH01003361 PCG66482.1 AGBW02008204 OWR54033.1 CCAG010015909 EZ422122 ADD18321.1 PYGN01000482 PSN45583.1 JXJN01015380 NEVH01020333 PNF21814.1 GBYB01000236 JAG70003.1 KK107148 QOIP01000009 EZA57441.1 RLU18392.1 NNAY01000950 OXU25762.1 KQ414646 KOC66509.1 GEZM01068162 JAV67125.1 LJIG01002384 KRT84556.1 CH480822 EDW55767.1 KQ434822 KZC07085.1 LJIG01022477 KRT80365.1 GL445235 EFN89997.1 GEZM01068163 JAV67124.1 AE014134 AY118739 AAF53808.1 AAM50599.1 GEZM01068164 JAV67123.1 GL762326 EFZ21688.1 CM000361 CM002910 EDX05518.1 KMY91022.1 CM000158 EDW90423.1 CH902620 EDV32037.1 KA648150 AFP62779.1 KZ288326 PBC27976.1 KPU73741.1 OUUW01000010 SPP85470.1 CH954179 EDV54647.1 GFDF01010660 JAV03424.1 CH379061 EAL33320.2 CH479195 EDW27305.1 CH940654 EDW57522.1 GFDF01010661 JAV03423.1 CP012523 ALC39496.1 KQ977279 KYN04265.1 ADTU01022920 GEZM01068165 JAV67122.1 CH916368 EDW03195.1 LBMM01005007 KMQ91920.1 GL436993 EFN71286.1 LSMT01000407 PFX18501.1 KQ980934 KYN11327.1 GAMC01003659 JAC02897.1 RCHS01001525 RMX53074.1 GL888719 EGI58276.1 AJVK01002574 GAKP01006877 JAC52075.1 CH963913 EDW77253.2 GDIP01180091 JAJ43311.1 GDIP01210041 JAJ13361.1 GDIQ01078458 JAN16279.1 GDIQ01171474 JAK80251.1 GDIQ01080352 JAN14385.1 AAZX01007538 GDIP01232447 JAI90954.1 GDIP01174795 JAJ48607.1 GDIQ01243453 JAK08272.1 GDIQ01191518 JAK60207.1 GDIP01153804 JAJ69598.1 GDIQ01255001 JAJ96723.1 KQ981382 KYN42139.1 GDIQ01036282 JAN58455.1 DS231838 EDS33886.1 GDIP01053572 JAM50143.1 LRGB01000024 KZS21585.1 GDIQ01088256 JAN06481.1 JRES01001524 KNC22245.1 KQ976464 KYM84429.1 GFDL01007447 JAV27598.1 KQ982652 KYQ52807.1 DS235430 EEB15674.1 GG666603 EEN50255.1 GDIP01222792 JAJ00610.1 GG666630 EEN47488.1 EDV32040.2 GBXI01002724 JAD11568.1 AERX01000001 AERX01000002 GDIQ01211206 JAK40519.1
KPJ07352.1 NWSH01003361 PCG66482.1 AGBW02008204 OWR54033.1 CCAG010015909 EZ422122 ADD18321.1 PYGN01000482 PSN45583.1 JXJN01015380 NEVH01020333 PNF21814.1 GBYB01000236 JAG70003.1 KK107148 QOIP01000009 EZA57441.1 RLU18392.1 NNAY01000950 OXU25762.1 KQ414646 KOC66509.1 GEZM01068162 JAV67125.1 LJIG01002384 KRT84556.1 CH480822 EDW55767.1 KQ434822 KZC07085.1 LJIG01022477 KRT80365.1 GL445235 EFN89997.1 GEZM01068163 JAV67124.1 AE014134 AY118739 AAF53808.1 AAM50599.1 GEZM01068164 JAV67123.1 GL762326 EFZ21688.1 CM000361 CM002910 EDX05518.1 KMY91022.1 CM000158 EDW90423.1 CH902620 EDV32037.1 KA648150 AFP62779.1 KZ288326 PBC27976.1 KPU73741.1 OUUW01000010 SPP85470.1 CH954179 EDV54647.1 GFDF01010660 JAV03424.1 CH379061 EAL33320.2 CH479195 EDW27305.1 CH940654 EDW57522.1 GFDF01010661 JAV03423.1 CP012523 ALC39496.1 KQ977279 KYN04265.1 ADTU01022920 GEZM01068165 JAV67122.1 CH916368 EDW03195.1 LBMM01005007 KMQ91920.1 GL436993 EFN71286.1 LSMT01000407 PFX18501.1 KQ980934 KYN11327.1 GAMC01003659 JAC02897.1 RCHS01001525 RMX53074.1 GL888719 EGI58276.1 AJVK01002574 GAKP01006877 JAC52075.1 CH963913 EDW77253.2 GDIP01180091 JAJ43311.1 GDIP01210041 JAJ13361.1 GDIQ01078458 JAN16279.1 GDIQ01171474 JAK80251.1 GDIQ01080352 JAN14385.1 AAZX01007538 GDIP01232447 JAI90954.1 GDIP01174795 JAJ48607.1 GDIQ01243453 JAK08272.1 GDIQ01191518 JAK60207.1 GDIP01153804 JAJ69598.1 GDIQ01255001 JAJ96723.1 KQ981382 KYN42139.1 GDIQ01036282 JAN58455.1 DS231838 EDS33886.1 GDIP01053572 JAM50143.1 LRGB01000024 KZS21585.1 GDIQ01088256 JAN06481.1 JRES01001524 KNC22245.1 KQ976464 KYM84429.1 GFDL01007447 JAV27598.1 KQ982652 KYQ52807.1 DS235430 EEB15674.1 GG666603 EEN50255.1 GDIP01222792 JAJ00610.1 GG666630 EEN47488.1 EDV32040.2 GBXI01002724 JAD11568.1 AERX01000001 AERX01000002 GDIQ01211206 JAK40519.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000092444
+ More
UP000245037 UP000092445 UP000092460 UP000235965 UP000078200 UP000053097 UP000279307 UP000215335 UP000053825 UP000001292 UP000076502 UP000008237 UP000005203 UP000000803 UP000095301 UP000000304 UP000002282 UP000007801 UP000242457 UP000268350 UP000008711 UP000001819 UP000008744 UP000095300 UP000008792 UP000092553 UP000192221 UP000078542 UP000005205 UP000001070 UP000036403 UP000000311 UP000225706 UP000078492 UP000275408 UP000007755 UP000092462 UP000007798 UP000002358 UP000078541 UP000002320 UP000076858 UP000007635 UP000037069 UP000078540 UP000075809 UP000009046 UP000076408 UP000001554 UP000192220 UP000005207
UP000245037 UP000092445 UP000092460 UP000235965 UP000078200 UP000053097 UP000279307 UP000215335 UP000053825 UP000001292 UP000076502 UP000008237 UP000005203 UP000000803 UP000095301 UP000000304 UP000002282 UP000007801 UP000242457 UP000268350 UP000008711 UP000001819 UP000008744 UP000095300 UP000008792 UP000092553 UP000192221 UP000078542 UP000005205 UP000001070 UP000036403 UP000000311 UP000225706 UP000078492 UP000275408 UP000007755 UP000092462 UP000007798 UP000002358 UP000078541 UP000002320 UP000076858 UP000007635 UP000037069 UP000078540 UP000075809 UP000009046 UP000076408 UP000001554 UP000192220 UP000005207
Interpro
ProteinModelPortal
H9J269
I4DPG6
A0A194PVF5
A0A0N1I4S7
A0A2A4J515
A0A212FJW5
+ More
A0A1B0FNM7 D3TL26 A0A2P8YMX7 A0A1B0A518 A0A1B0BJG1 A0A2J7PZQ0 A0A0C9R086 A0A1A9V9K8 A0A026WP48 A0A232F570 A0A0L7R6Q5 A0A1Y1L567 A0A0T6BB49 B4I5W1 A0A154P563 A0A0T6AZS1 E2B2T2 A0A088AGP6 A0A1Y1L0L0 Q9VIV6 A0A1Y1L0E2 A0A1I8MI79 E9ICM1 B4Q9Y8 B4PAZ4 B3MN59 T1PJ57 A0A2A3E9X4 A0A0P8XFP8 A0A3B0KBE1 B3NM47 A0A1L8DAT2 Q29K58 B4GWX8 A0A1I8PRM5 B4M8J6 A0A1L8DAD0 A0A0M4E5Q9 A0A1I8PRR9 A0A1W4VG39 A0A195CUC9 A0A158NQ78 A0A1Y1L0D4 B4JCV6 A0A0J7NHD2 E2A5K3 A0A1I8M2J5 A0A1I8NUH6 A0A2B4RLM4 A0A195DER3 W8C7D1 A0A3M6UHR7 A0A1I8Q5P3 F4X5I6 A0A1B0D1D1 A0A034WD54 B4MYR4 A0A1I8MFU3 A0A0P5C3X5 A0A0N7ZUY7 A0A0P6EDN2 A0A0P5LD38 A0A0P6DDT0 K7IV53 A0A0P4Y577 A0A0P5C827 A0A0P5GS02 A0A0P5KFN9 A0A0P5DRG9 A0A0P5H4F7 A0A195FQE5 A0A0P6H1A2 B0W4P1 A0A0P5YTT5 A0A162SSA6 A0A0P6CRI3 G3PVB0 A0A0L0BQ83 A0A195BJ86 A0A1Q3FJ93 A0A151WYH1 E0VQL8 A0A1I8P8E6 A0A182YMQ6 C3ZBL8 A0A0P4YTT0 C3ZIX2 B3MN62 A0A0A1XLV1 A0A2I4CKG6 I3J2E7 A0A0P5J495
A0A1B0FNM7 D3TL26 A0A2P8YMX7 A0A1B0A518 A0A1B0BJG1 A0A2J7PZQ0 A0A0C9R086 A0A1A9V9K8 A0A026WP48 A0A232F570 A0A0L7R6Q5 A0A1Y1L567 A0A0T6BB49 B4I5W1 A0A154P563 A0A0T6AZS1 E2B2T2 A0A088AGP6 A0A1Y1L0L0 Q9VIV6 A0A1Y1L0E2 A0A1I8MI79 E9ICM1 B4Q9Y8 B4PAZ4 B3MN59 T1PJ57 A0A2A3E9X4 A0A0P8XFP8 A0A3B0KBE1 B3NM47 A0A1L8DAT2 Q29K58 B4GWX8 A0A1I8PRM5 B4M8J6 A0A1L8DAD0 A0A0M4E5Q9 A0A1I8PRR9 A0A1W4VG39 A0A195CUC9 A0A158NQ78 A0A1Y1L0D4 B4JCV6 A0A0J7NHD2 E2A5K3 A0A1I8M2J5 A0A1I8NUH6 A0A2B4RLM4 A0A195DER3 W8C7D1 A0A3M6UHR7 A0A1I8Q5P3 F4X5I6 A0A1B0D1D1 A0A034WD54 B4MYR4 A0A1I8MFU3 A0A0P5C3X5 A0A0N7ZUY7 A0A0P6EDN2 A0A0P5LD38 A0A0P6DDT0 K7IV53 A0A0P4Y577 A0A0P5C827 A0A0P5GS02 A0A0P5KFN9 A0A0P5DRG9 A0A0P5H4F7 A0A195FQE5 A0A0P6H1A2 B0W4P1 A0A0P5YTT5 A0A162SSA6 A0A0P6CRI3 G3PVB0 A0A0L0BQ83 A0A195BJ86 A0A1Q3FJ93 A0A151WYH1 E0VQL8 A0A1I8P8E6 A0A182YMQ6 C3ZBL8 A0A0P4YTT0 C3ZIX2 B3MN62 A0A0A1XLV1 A0A2I4CKG6 I3J2E7 A0A0P5J495
PDB
3QSJ
E-value=0.00525264,
Score=90
Ontologies
GO
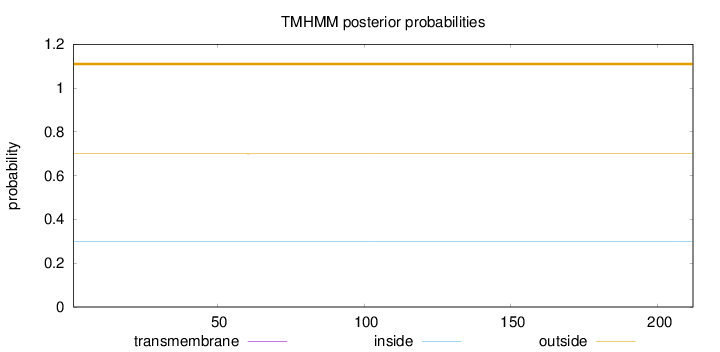
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00458
Exp number, first 60 AAs:
0.000710000000000001
Total prob of N-in:
0.30048
outside
1 - 212
Population Genetic Test Statistics
Pi
207.781343
Theta
143.580663
Tajima's D
-0.774582
CLR
0.51899
CSRT
0.184140792960352
Interpretation
Uncertain
