Gene
KWMTBOMO03007 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003607
Annotation
PREDICTED:_STE20/SPS1-related_proline-alanine-rich_protein_kinase_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.066 Nuclear Reliability : 1.711
Sequence
CDS
ATGGCTACCTCCGGTAACTCGTGGCCTAACAGCCAGGATGATTATGAACTGATGGAAGTGATCGGGGTCGGGGCCACGGCCGTCGTGCACGCCGCCTTCTGCCGCCCGCGGGGAGAGAAGTGCGCCATCAAGCGTATCAACCTCGAGAAATGGAACACCTCCATGGATGAACTGCTCAAAGAGATTCAGGCCATGTCAAGCTGCAACCACGAAAATGTCGTCACCTATTACACCAGCTTTGTGGTAAAAGAAGAACTGTGGCTCGTTCTGAGACTTCTCGAAGGAGGAAGTCTTCTTGACATCATCAAACATAAGATGAGGATCTCCAACTGTAAACATGGAGTATTTGATGAAGCGACCATCGCCACGGTTCTCAAGGAGGTGCTTAAAGGATTAGAATACTTCCATCAGAACGGGCAAATTCACAGGGATATTAAAGCAGGAAATATCCTCCTCGGTGATGATGGAACCGTCCAGATCGCCGACTTCGGTGTGTCTGCATGGCTCGCGACCGGCCGGGACTTGTCCAGACAGAAGGTGCGACATACATTCGTCGGGACGCCTTGTTGGATGGCTCCCGAGGTCATGGAACAAAATCACGGTTATGACTTCAAAGCGGATATCTGGTCGTTCGGCATCACCGCGATCGAGATGGCCACCGGCACCGCCCCGTACCACAAGTACCCACCGATGAAGGTACTCATGCTCACGTTGCAGAATGACCCGCCAACACTCGACACCGGCGCCGAGGAAAAAGATCAGTACAAAGCGTACGGTAAGACTTTCAGGAAAATGATATCGGAGTGTCTCCAGAAAGATCCAACGAAACGACCTTCGGCGCCGGAACTGCTCAAGCACTCGTTTTTCAAGAAAGCAAAGGATCGAAAGTATCTCGTGCAGACGCTCGTCTCGAACGGCCCGAGCATGGAGACGCGCGTGCACAAGGCCAGCAAGCGACAACCTGGAGCCTCCGGTCGTCTGCATCGCATGGAGACCGGGGAGTGGGTGTGGGAGAGCGACGACGAAGAGGACGAGGACGATGACGCCGCCAGGGACCGACCCATGAACACCCTCGCACGTGCAGACTCCTCCGACTCCGAGGGTGAGGAGCTTGGCGAAGGTCGCGCTGGTTTCACGATCGGTTCGCTCACTGGTGGCGAGGCTCCACAGATCGACCTCGTATTGCGCATGCGTAATTCTAAGCGCGAGCTGAACGACATCCGGTTCGAATTCTCGGCCGGCAAGGATTCCGCCGACGGCATCGCCACGGAGCTGGTGGGTGCGGGGCTGGTGGAGGCGCGGGACGCCGCGTGCATCGCCCGACACCTCCAGCGGCTGGTGGACGCGCGCCTGTTCCCCGGCTCGGCGCCCCCGCCGCGCTCGCTCACCTTCCGGCTGGACTCTGCGGACCCGGCCGCGCCCTCCGACGAGAGCACGCTGCTCGGGTTCGCGCAGATCTCTGTAGTGGACTAA
Protein
MATSGNSWPNSQDDYELMEVIGVGATAVVHAAFCRPRGEKCAIKRINLEKWNTSMDELLKEIQAMSSCNHENVVTYYTSFVVKEELWLVLRLLEGGSLLDIIKHKMRISNCKHGVFDEATIATVLKEVLKGLEYFHQNGQIHRDIKAGNILLGDDGTVQIADFGVSAWLATGRDLSRQKVRHTFVGTPCWMAPEVMEQNHGYDFKADIWSFGITAIEMATGTAPYHKYPPMKVLMLTLQNDPPTLDTGAEEKDQYKAYGKTFRKMISECLQKDPTKRPSAPELLKHSFFKKAKDRKYLVQTLVSNGPSMETRVHKASKRQPGASGRLHRMETGEWVWESDDEEDEDDDAARDRPMNTLARADSSDSEGEELGEGRAGFTIGSLTGGEAPQIDLVLRMRNSKRELNDIRFEFSAGKDSADGIATELVGAGLVEARDAACIARHLQRLVDARLFPGSAPPPRSLTFRLDSADPAAPSDESTLLGFAQISVVD
Summary
Uniprot
H9J270
A0A194PQT8
A0A0N0PAY3
A0A2A4J4X8
A0A2H1VPW2
A0A0L7LI17
+ More
U5EKR3 D6WLE3 A0A1L8DLL4 A0A1L8DLU7 A0A1Y1NIE1 A0A1L8DLV8 A0A2A4JWU6 A0A1L8DLJ6 A0A1W4WDG5 A0A212FLN9 Q16PN5 A0A1B0C9U0 A0A182H2D3 T1D3X3 A0A1Q3F315 A0A1Q3F2S7 A0A1B0GMS4 B0X7M4 A0A2H1VIF6 E0VXD4 A0A212FJX7 A0A336LFN7 A0A2J7Q4Z4 A0A1Y1NIA2 A0A224XBR4 A0A0P9AQE8 A0A067RM82 A0A1B6C1U1 A0A1B6CN80 A0A069DUR6 T1E1V7 A0A0L0BVU0 A0A0P8XZ01 B3NZ48 A0A1W4UJW7 B4I279 A0A1W4UJZ7 Q9VE62 A0A3B0KRX9 Q7KSD3 B4NH29 A0A026W0F9 K7J7Z3 A0A0N0BFH5 E2BNX1 A0A151K001 V9IEA4 A0A2M4A1J2 A0A2M4A149 W5JER2 Q299M8 A0A0M4ENL8 A0A151I5W0 A0A2M3ZGC8 A0A182FFT5 A0A182SAH6 A0A2M4BIQ0 A0A0A9VPI4 A0A0K8T5R0 V9IBY0 A0A182YNJ9 A0A0K8T623 A0A084WLL3 A0A0L7R0H7 A0A182KLE3 A0A3L8DNV7 A0A2A3EP44 A0A087ZP17 A0A151XH10 A0A182TYY8 A0A151IG27 A0A182V1R9 A0A158P1A3 A0A182X1B4 A0A182M8Y5 A0A182PHU3 A0A182WIB6 Q7QI85 A0A182K3Y0 A0A182R2H2 A0A182QW69 A0A182JDZ4 A0A310SIQ9 A0A182N7P4 A0A1J1HR40 B4QUD8 N6T2Q0 A0A1D2M8Z0 F4WA32 T1HV69 A0A1S3D4V7 A0A3Q0J2A7 A0A154NZJ4 J9JZ38
U5EKR3 D6WLE3 A0A1L8DLL4 A0A1L8DLU7 A0A1Y1NIE1 A0A1L8DLV8 A0A2A4JWU6 A0A1L8DLJ6 A0A1W4WDG5 A0A212FLN9 Q16PN5 A0A1B0C9U0 A0A182H2D3 T1D3X3 A0A1Q3F315 A0A1Q3F2S7 A0A1B0GMS4 B0X7M4 A0A2H1VIF6 E0VXD4 A0A212FJX7 A0A336LFN7 A0A2J7Q4Z4 A0A1Y1NIA2 A0A224XBR4 A0A0P9AQE8 A0A067RM82 A0A1B6C1U1 A0A1B6CN80 A0A069DUR6 T1E1V7 A0A0L0BVU0 A0A0P8XZ01 B3NZ48 A0A1W4UJW7 B4I279 A0A1W4UJZ7 Q9VE62 A0A3B0KRX9 Q7KSD3 B4NH29 A0A026W0F9 K7J7Z3 A0A0N0BFH5 E2BNX1 A0A151K001 V9IEA4 A0A2M4A1J2 A0A2M4A149 W5JER2 Q299M8 A0A0M4ENL8 A0A151I5W0 A0A2M3ZGC8 A0A182FFT5 A0A182SAH6 A0A2M4BIQ0 A0A0A9VPI4 A0A0K8T5R0 V9IBY0 A0A182YNJ9 A0A0K8T623 A0A084WLL3 A0A0L7R0H7 A0A182KLE3 A0A3L8DNV7 A0A2A3EP44 A0A087ZP17 A0A151XH10 A0A182TYY8 A0A151IG27 A0A182V1R9 A0A158P1A3 A0A182X1B4 A0A182M8Y5 A0A182PHU3 A0A182WIB6 Q7QI85 A0A182K3Y0 A0A182R2H2 A0A182QW69 A0A182JDZ4 A0A310SIQ9 A0A182N7P4 A0A1J1HR40 B4QUD8 N6T2Q0 A0A1D2M8Z0 F4WA32 T1HV69 A0A1S3D4V7 A0A3Q0J2A7 A0A154NZJ4 J9JZ38
Pubmed
19121390
26354079
26227816
18362917
19820115
28004739
+ More
22118469 17510324 26483478 24330624 20566863 17994087 24845553 26334808 26108605 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 20075255 20798317 20920257 23761445 15632085 25401762 26823975 25244985 24438588 20966253 30249741 21347285 12364791 14747013 17210077 23537049 27289101 21719571
22118469 17510324 26483478 24330624 20566863 17994087 24845553 26334808 26108605 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 20075255 20798317 20920257 23761445 15632085 25401762 26823975 25244985 24438588 20966253 30249741 21347285 12364791 14747013 17210077 23537049 27289101 21719571
EMBL
BABH01007424
KQ459597
KPI95109.1
KQ461187
KQ459677
KPJ07351.1
+ More
KPJ20479.1 NWSH01003361 PCG66480.1 ODYU01003719 SOQ42850.1 JTDY01001024 KOB75097.1 GANO01001709 JAB58162.1 KQ971343 EFA04100.2 GFDF01006738 JAV07346.1 GFDF01006739 JAV07345.1 GEZM01001649 JAV97621.1 GFDF01006740 JAV07344.1 NWSH01000478 PCG76114.1 GFDF01006741 JAV07343.1 AGBW02007730 OWR54652.1 CH477776 EAT36321.1 AJWK01002765 JXUM01105128 JXUM01105129 JXUM01105130 JXUM01105131 GALA01001193 JAA93659.1 GFDL01013102 JAV21943.1 GFDL01013185 JAV21860.1 AJVK01026458 AJVK01026459 DS232455 EDS42036.1 ODYU01002745 SOQ40627.1 DS235830 EEB18040.1 AGBW02008204 OWR54034.1 UFQS01003329 UFQT01003329 SSX15513.1 SSX34879.1 NEVH01018373 PNF23646.1 GEZM01001647 JAV97623.1 GFTR01006560 JAW09866.1 CH902617 KPU79838.1 KK852543 KDR21690.1 GEDC01029816 JAS07482.1 GEDC01022382 JAS14916.1 GBGD01001333 JAC87556.1 GALA01001720 JAA93132.1 JRES01001362 KNC23354.1 KPU79836.1 KPU79839.1 CH954181 EDV48590.1 CH480820 EDW54636.1 AF006640 AAD01275.1 OUUW01000013 SPP87991.1 AE014297 BT003198 BT021945 AAF55567.2 AAN13778.1 AAO24953.1 AAX51650.1 ACL83535.1 ACL83536.1 ACL83537.1 AHN57394.1 ALI30588.1 ALI30589.1 CH964272 EDW84526.2 KK107503 EZA49553.1 KQ435798 KOX73362.1 GL449511 EFN82563.1 KQ981301 KYN43053.1 JR039298 AEY58624.1 GGFK01001187 MBW34508.1 GGFK01001196 MBW34517.1 ADMH02001359 ETN62842.1 CM000070 EAL27673.4 CP012526 ALC46314.1 KQ976402 KYM92288.1 GGFM01006759 MBW27510.1 GGFJ01003798 MBW52939.1 GBHO01045182 GBRD01004952 GBRD01004951 GDHC01006505 JAF98421.1 JAG60869.1 JAQ12124.1 GBRD01004949 GDHC01021265 JAG60872.1 JAP97363.1 JR039297 AEY58623.1 GBRD01004950 JAG60871.1 ATLV01024255 KE525350 KFB51107.1 KQ414670 KOC64347.1 QOIP01000006 RLU21966.1 KZ288204 PBC33284.1 KQ982138 KYQ59686.1 KQ977743 KYN00171.1 ADTU01006165 ADTU01006166 ADTU01006167 ADTU01006168 ADTU01006169 ADTU01006170 AXCM01007456 AAAB01008807 EAA04686.4 AXCN02000582 KQ762329 OAD55960.1 CVRI01000019 CRK90511.1 CM000364 EDX12438.1 APGK01046715 KB741067 ENN74404.1 LJIJ01002640 ODM89438.1 GL888037 EGI68962.1 ACPB03007618 KQ434781 KZC04504.1 ABLF02035510
KPJ20479.1 NWSH01003361 PCG66480.1 ODYU01003719 SOQ42850.1 JTDY01001024 KOB75097.1 GANO01001709 JAB58162.1 KQ971343 EFA04100.2 GFDF01006738 JAV07346.1 GFDF01006739 JAV07345.1 GEZM01001649 JAV97621.1 GFDF01006740 JAV07344.1 NWSH01000478 PCG76114.1 GFDF01006741 JAV07343.1 AGBW02007730 OWR54652.1 CH477776 EAT36321.1 AJWK01002765 JXUM01105128 JXUM01105129 JXUM01105130 JXUM01105131 GALA01001193 JAA93659.1 GFDL01013102 JAV21943.1 GFDL01013185 JAV21860.1 AJVK01026458 AJVK01026459 DS232455 EDS42036.1 ODYU01002745 SOQ40627.1 DS235830 EEB18040.1 AGBW02008204 OWR54034.1 UFQS01003329 UFQT01003329 SSX15513.1 SSX34879.1 NEVH01018373 PNF23646.1 GEZM01001647 JAV97623.1 GFTR01006560 JAW09866.1 CH902617 KPU79838.1 KK852543 KDR21690.1 GEDC01029816 JAS07482.1 GEDC01022382 JAS14916.1 GBGD01001333 JAC87556.1 GALA01001720 JAA93132.1 JRES01001362 KNC23354.1 KPU79836.1 KPU79839.1 CH954181 EDV48590.1 CH480820 EDW54636.1 AF006640 AAD01275.1 OUUW01000013 SPP87991.1 AE014297 BT003198 BT021945 AAF55567.2 AAN13778.1 AAO24953.1 AAX51650.1 ACL83535.1 ACL83536.1 ACL83537.1 AHN57394.1 ALI30588.1 ALI30589.1 CH964272 EDW84526.2 KK107503 EZA49553.1 KQ435798 KOX73362.1 GL449511 EFN82563.1 KQ981301 KYN43053.1 JR039298 AEY58624.1 GGFK01001187 MBW34508.1 GGFK01001196 MBW34517.1 ADMH02001359 ETN62842.1 CM000070 EAL27673.4 CP012526 ALC46314.1 KQ976402 KYM92288.1 GGFM01006759 MBW27510.1 GGFJ01003798 MBW52939.1 GBHO01045182 GBRD01004952 GBRD01004951 GDHC01006505 JAF98421.1 JAG60869.1 JAQ12124.1 GBRD01004949 GDHC01021265 JAG60872.1 JAP97363.1 JR039297 AEY58623.1 GBRD01004950 JAG60871.1 ATLV01024255 KE525350 KFB51107.1 KQ414670 KOC64347.1 QOIP01000006 RLU21966.1 KZ288204 PBC33284.1 KQ982138 KYQ59686.1 KQ977743 KYN00171.1 ADTU01006165 ADTU01006166 ADTU01006167 ADTU01006168 ADTU01006169 ADTU01006170 AXCM01007456 AAAB01008807 EAA04686.4 AXCN02000582 KQ762329 OAD55960.1 CVRI01000019 CRK90511.1 CM000364 EDX12438.1 APGK01046715 KB741067 ENN74404.1 LJIJ01002640 ODM89438.1 GL888037 EGI68962.1 ACPB03007618 KQ434781 KZC04504.1 ABLF02035510
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000037510
UP000007266
+ More
UP000192223 UP000007151 UP000008820 UP000092461 UP000069940 UP000092462 UP000002320 UP000009046 UP000235965 UP000007801 UP000027135 UP000037069 UP000008711 UP000192221 UP000001292 UP000268350 UP000000803 UP000007798 UP000053097 UP000002358 UP000053105 UP000008237 UP000078541 UP000000673 UP000001819 UP000092553 UP000078540 UP000069272 UP000075901 UP000076408 UP000030765 UP000053825 UP000075882 UP000279307 UP000242457 UP000005203 UP000075809 UP000075902 UP000078542 UP000075903 UP000005205 UP000076407 UP000075883 UP000075885 UP000075920 UP000007062 UP000075881 UP000075900 UP000075886 UP000075880 UP000075884 UP000183832 UP000000304 UP000019118 UP000094527 UP000007755 UP000015103 UP000079169 UP000076502 UP000007819
UP000192223 UP000007151 UP000008820 UP000092461 UP000069940 UP000092462 UP000002320 UP000009046 UP000235965 UP000007801 UP000027135 UP000037069 UP000008711 UP000192221 UP000001292 UP000268350 UP000000803 UP000007798 UP000053097 UP000002358 UP000053105 UP000008237 UP000078541 UP000000673 UP000001819 UP000092553 UP000078540 UP000069272 UP000075901 UP000076408 UP000030765 UP000053825 UP000075882 UP000279307 UP000242457 UP000005203 UP000075809 UP000075902 UP000078542 UP000075903 UP000005205 UP000076407 UP000075883 UP000075885 UP000075920 UP000007062 UP000075881 UP000075900 UP000075886 UP000075880 UP000075884 UP000183832 UP000000304 UP000019118 UP000094527 UP000007755 UP000015103 UP000079169 UP000076502 UP000007819
Interpro
Gene 3D
ProteinModelPortal
H9J270
A0A194PQT8
A0A0N0PAY3
A0A2A4J4X8
A0A2H1VPW2
A0A0L7LI17
+ More
U5EKR3 D6WLE3 A0A1L8DLL4 A0A1L8DLU7 A0A1Y1NIE1 A0A1L8DLV8 A0A2A4JWU6 A0A1L8DLJ6 A0A1W4WDG5 A0A212FLN9 Q16PN5 A0A1B0C9U0 A0A182H2D3 T1D3X3 A0A1Q3F315 A0A1Q3F2S7 A0A1B0GMS4 B0X7M4 A0A2H1VIF6 E0VXD4 A0A212FJX7 A0A336LFN7 A0A2J7Q4Z4 A0A1Y1NIA2 A0A224XBR4 A0A0P9AQE8 A0A067RM82 A0A1B6C1U1 A0A1B6CN80 A0A069DUR6 T1E1V7 A0A0L0BVU0 A0A0P8XZ01 B3NZ48 A0A1W4UJW7 B4I279 A0A1W4UJZ7 Q9VE62 A0A3B0KRX9 Q7KSD3 B4NH29 A0A026W0F9 K7J7Z3 A0A0N0BFH5 E2BNX1 A0A151K001 V9IEA4 A0A2M4A1J2 A0A2M4A149 W5JER2 Q299M8 A0A0M4ENL8 A0A151I5W0 A0A2M3ZGC8 A0A182FFT5 A0A182SAH6 A0A2M4BIQ0 A0A0A9VPI4 A0A0K8T5R0 V9IBY0 A0A182YNJ9 A0A0K8T623 A0A084WLL3 A0A0L7R0H7 A0A182KLE3 A0A3L8DNV7 A0A2A3EP44 A0A087ZP17 A0A151XH10 A0A182TYY8 A0A151IG27 A0A182V1R9 A0A158P1A3 A0A182X1B4 A0A182M8Y5 A0A182PHU3 A0A182WIB6 Q7QI85 A0A182K3Y0 A0A182R2H2 A0A182QW69 A0A182JDZ4 A0A310SIQ9 A0A182N7P4 A0A1J1HR40 B4QUD8 N6T2Q0 A0A1D2M8Z0 F4WA32 T1HV69 A0A1S3D4V7 A0A3Q0J2A7 A0A154NZJ4 J9JZ38
U5EKR3 D6WLE3 A0A1L8DLL4 A0A1L8DLU7 A0A1Y1NIE1 A0A1L8DLV8 A0A2A4JWU6 A0A1L8DLJ6 A0A1W4WDG5 A0A212FLN9 Q16PN5 A0A1B0C9U0 A0A182H2D3 T1D3X3 A0A1Q3F315 A0A1Q3F2S7 A0A1B0GMS4 B0X7M4 A0A2H1VIF6 E0VXD4 A0A212FJX7 A0A336LFN7 A0A2J7Q4Z4 A0A1Y1NIA2 A0A224XBR4 A0A0P9AQE8 A0A067RM82 A0A1B6C1U1 A0A1B6CN80 A0A069DUR6 T1E1V7 A0A0L0BVU0 A0A0P8XZ01 B3NZ48 A0A1W4UJW7 B4I279 A0A1W4UJZ7 Q9VE62 A0A3B0KRX9 Q7KSD3 B4NH29 A0A026W0F9 K7J7Z3 A0A0N0BFH5 E2BNX1 A0A151K001 V9IEA4 A0A2M4A1J2 A0A2M4A149 W5JER2 Q299M8 A0A0M4ENL8 A0A151I5W0 A0A2M3ZGC8 A0A182FFT5 A0A182SAH6 A0A2M4BIQ0 A0A0A9VPI4 A0A0K8T5R0 V9IBY0 A0A182YNJ9 A0A0K8T623 A0A084WLL3 A0A0L7R0H7 A0A182KLE3 A0A3L8DNV7 A0A2A3EP44 A0A087ZP17 A0A151XH10 A0A182TYY8 A0A151IG27 A0A182V1R9 A0A158P1A3 A0A182X1B4 A0A182M8Y5 A0A182PHU3 A0A182WIB6 Q7QI85 A0A182K3Y0 A0A182R2H2 A0A182QW69 A0A182JDZ4 A0A310SIQ9 A0A182N7P4 A0A1J1HR40 B4QUD8 N6T2Q0 A0A1D2M8Z0 F4WA32 T1HV69 A0A1S3D4V7 A0A3Q0J2A7 A0A154NZJ4 J9JZ38
Ontologies
GO
PANTHER
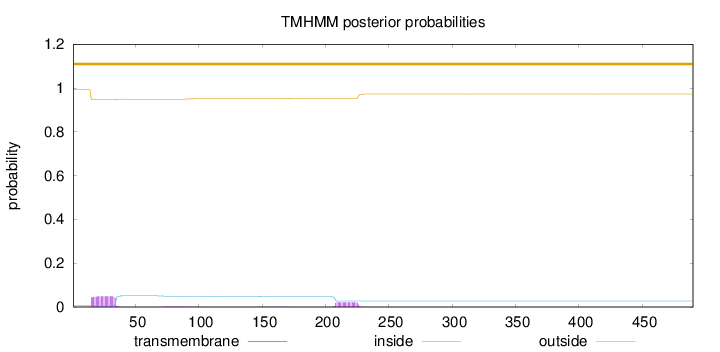
Topology
Length:
490
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.42066
Exp number, first 60 AAs:
0.96116
Total prob of N-in:
0.00724
outside
1 - 490
Population Genetic Test Statistics
Pi
230.259266
Theta
237.449391
Tajima's D
0.103323
CLR
30.74195
CSRT
0.394380280985951
Interpretation
Uncertain
