Gene
KWMTBOMO03006
Pre Gene Modal
BGIBMGA003588
Annotation
FERM_domain-containing_protein_5_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.365 Nuclear Reliability : 1.148
Sequence
CDS
ATGTTTAAAAGCCGTGGAGATATTGTTTATAAATGCACTGTGCGACTTTTAGAGGATACTGAAATTTTAGAATGTGAATTTCATCCTAGTTTTAAGGGTAAATCTCTGCTAGAACATGTGTGTCAACAATTGAATCTCACAGAGTCTGACTACTTTGGTTTAAGATATGTGGACGCAAGTGGACAGAGACACTGGCTACACATGGCTAAACTCATCCTCAAACAAGTCAAAGATGCTTCACCTATATTGTTCAGTTTTCGAGTGAAGTTCTATCCACCTAATCCTTTACTACTAAAGGAAGATGTGACTAGATTCCAAATATATTTGCAATTGAAGCGGGATCTACTTCATGGCAGGTTGTACTGTACAGCCAATGAAGCTGCCATGCTTGGAGCTCTCATCATTCAAATTGAATTGGGAGACTATGATTCAACTATCCACATTGGTAATTATGTGAGCGAAATGAGATTGTTATTGAGACAAACAGATGCAATTGAAGGTAGGATACAGGAACTACATAGGGAACCATTGGAAGGCACACCCATTGGTGGTGGAGGAGTTAAGGGTATGTCAACTGAAGAAGCCGAAAGAACATTCCTAAAGATTGCTTGCCAACTTGATACTTATGGAGTGGACCCGCATCCAGTTAAGGATCACAGAGGAAACCAGTTATATTTAGGTATCAACCACAGTGGCATATTAACATTTCAAGGCAGTAGGAAAACAAATCATTTCAAATGGTCAGAAGTTCACAAGATCAACTTTGAGGGGAGAATGTTTATTGTTCACTTGAATTACCCAGAGAAAAAACACACGGTCGGCTTCAAGTGCCCGAGCGGGGCGGCTTGTCGTCACGTCTGGTGCTGTGCCATAGAGCAGATGCTGTTCTTCACTTTGTCGTCATCGTCGGAAGCGTGCGTGTACTCTGGCGGGGGACTGTTCTCGTGGGGAACGAAATTTAAGTACGTGGGGCGGACAGAGAGGGAGATCCTCGAGAGGGACGGCCTCCTGGCACCGAGGGAGCCTCAGGAAGAAGGCTCCAGTGCTGGAGGAAAGAGGAAAGCTTCGAGCGTACCAGCGACTCCGTCTACGCCGATGACTGGGGACTTTGGGTACAGCAGTCTGCCGCGCTCGACGCACAGCGCGCCGCTGGAGGAGAGCGGGGACGGCGCCGCGCACCTCGCGCTGGCGTGTTGCGAGCGCGCACCCGCCGACAAGCCCGCCGTATGTCCGTCCGAGTACGGGATGCGGGACTCGTTCGAGCACTCGTCGTCCGAGTCTGGCGTGACGTCACAGACGGGTCCGTCTCAGGGCGAGGAGTGGAGCCGGCCCCCCACCGCGGCGCCCCCGCCCCCCGCGCCGCGCCCCTTCAGCCTCGTGCGCGCCTTCGTGCCGTCGTTCCTGTTCGTGTGGCTGTCGCTAGGTATGTTCGCACTCCTGCTGTTCGAGACGGACGTGCGTTTTCTGCGCGCCGTCAAGCACAAGCCGGAAATGGTGGCGCTGCGCCGGTACTACTACGCGCCCCTCAAGGAATACGTCAAGAGAAAGGTCGTTGAACTGTTTTAA
Protein
MFKSRGDIVYKCTVRLLEDTEILECEFHPSFKGKSLLEHVCQQLNLTESDYFGLRYVDASGQRHWLHMAKLILKQVKDASPILFSFRVKFYPPNPLLLKEDVTRFQIYLQLKRDLLHGRLYCTANEAAMLGALIIQIELGDYDSTIHIGNYVSEMRLLLRQTDAIEGRIQELHREPLEGTPIGGGGVKGMSTEEAERTFLKIACQLDTYGVDPHPVKDHRGNQLYLGINHSGILTFQGSRKTNHFKWSEVHKINFEGRMFIVHLNYPEKKHTVGFKCPSGAACRHVWCCAIEQMLFFTLSSSSEACVYSGGGLFSWGTKFKYVGRTEREILERDGLLAPREPQEEGSSAGGKRKASSVPATPSTPMTGDFGYSSLPRSTHSAPLEESGDGAAHLALACCERAPADKPAVCPSEYGMRDSFEHSSSESGVTSQTGPSQGEEWSRPPTAAPPPPAPRPFSLVRAFVPSFLFVWLSLGMFALLLFETDVRFLRAVKHKPEMVALRRYYYAPLKEYVKRKVVELF
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
H9J251
A0A2H1VPU7
A0A194PQ68
A0A2A4J485
A0A2A4J3P0
A0A2A4J2V8
+ More
A0A212FJW6 A0A0N0PAM2 D6W6N9 A0A1B6CYS5 A0A1Y1KQS4 A0A2J7R3Q3 N6TSX7 A0A2J7R3S6 A0A2J7R3S0 A0A2J7R3R3 A0A1W4XG86 A0A1W4XRA3 A0A1B0CHP0 A0A1B6L3F3 B0WPV3 A0A087ZS10 A0A154PEU2 B4KHT0 B3MPS0 A0A0C9QUZ1 B4JPY4 B4LVD7 B4NZA6 Q17BE5 B4HWL5 A0A0M3QTP3 A0A182GDD2 Q9VKY7 B3N9M1 B4Q970 A0A0C9PSY0 A0A1W4USX9 A0A2S2PEH3 A0A2S2NL20 A0A336MDB5 A0A0A9XWI4 A0A182MAW3 J9JZI4 A0A182R2J6 A0A084VJ89 A0A2S2QW22 A0A182WA08 A0A182UQW5 Q7Q844 Q29CW6 B4G7K2 A0A182Y9A6 A0A182NT53 B4NLV3 A0A026WRH3 A0A195E5K1 E0VQN4 A0A182IMJ2 E2ADQ1 A0A151I8X7 A0A182LK09 A0A182IB29 A0A158NR13 A0A1B0AV55 A0A3L8DW25 A0A1A9ZM10 A0A1A9Y5P4 A0A3B0JLC9 A0A1A9WJ43 A0A023F1K5 A0A1A9UTA3 A0A3B0JMU0 A0A2R5LNG6 A0A1I8Q1V9 A0A1J1ILS3 A0A1Z5L583 A0A131YWU2 A0A182F2U3 A0A195EUB8 L7M2I0 A0A2A3E434 W8B6G4 A0A1B0FPV8 Q86P10 A0A0R3NWK1 A0A1D2MN79
A0A212FJW6 A0A0N0PAM2 D6W6N9 A0A1B6CYS5 A0A1Y1KQS4 A0A2J7R3Q3 N6TSX7 A0A2J7R3S6 A0A2J7R3S0 A0A2J7R3R3 A0A1W4XG86 A0A1W4XRA3 A0A1B0CHP0 A0A1B6L3F3 B0WPV3 A0A087ZS10 A0A154PEU2 B4KHT0 B3MPS0 A0A0C9QUZ1 B4JPY4 B4LVD7 B4NZA6 Q17BE5 B4HWL5 A0A0M3QTP3 A0A182GDD2 Q9VKY7 B3N9M1 B4Q970 A0A0C9PSY0 A0A1W4USX9 A0A2S2PEH3 A0A2S2NL20 A0A336MDB5 A0A0A9XWI4 A0A182MAW3 J9JZI4 A0A182R2J6 A0A084VJ89 A0A2S2QW22 A0A182WA08 A0A182UQW5 Q7Q844 Q29CW6 B4G7K2 A0A182Y9A6 A0A182NT53 B4NLV3 A0A026WRH3 A0A195E5K1 E0VQN4 A0A182IMJ2 E2ADQ1 A0A151I8X7 A0A182LK09 A0A182IB29 A0A158NR13 A0A1B0AV55 A0A3L8DW25 A0A1A9ZM10 A0A1A9Y5P4 A0A3B0JLC9 A0A1A9WJ43 A0A023F1K5 A0A1A9UTA3 A0A3B0JMU0 A0A2R5LNG6 A0A1I8Q1V9 A0A1J1ILS3 A0A1Z5L583 A0A131YWU2 A0A182F2U3 A0A195EUB8 L7M2I0 A0A2A3E434 W8B6G4 A0A1B0FPV8 Q86P10 A0A0R3NWK1 A0A1D2MN79
Pubmed
19121390
26354079
22118469
18362917
19820115
28004739
+ More
23537049 17994087 17550304 17510324 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25401762 26823975 24438588 12364791 14747013 17210077 15632085 25244985 24508170 20566863 20798317 20966253 21347285 30249741 25474469 28528879 26830274 25576852 24495485 27289101
23537049 17994087 17550304 17510324 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25401762 26823975 24438588 12364791 14747013 17210077 15632085 25244985 24508170 20566863 20798317 20966253 21347285 30249741 25474469 28528879 26830274 25576852 24495485 27289101
EMBL
BABH01007424
BABH01007425
BABH01007426
ODYU01003719
SOQ42849.1
KQ459597
+ More
KPI95108.1 NWSH01003361 PCG66478.1 PCG66479.1 PCG66477.1 AGBW02008204 OWR54035.1 KQ461187 KPJ07350.1 KQ971307 EFA10977.1 GEDC01029751 GEDC01018823 JAS07547.1 JAS18475.1 GEZM01079030 JAV62530.1 NEVH01007818 PNF35471.1 APGK01020369 KB740193 KB631602 ENN81148.1 ERL84544.1 PNF35470.1 PNF35472.1 PNF35473.1 AJWK01012548 GEBQ01021816 JAT18161.1 DS232030 EDS32541.1 KQ434879 KZC09954.1 CH933807 EDW13367.1 CH902620 EDV32318.1 GBYB01004457 JAG74224.1 CH916372 EDV98964.1 CH940649 EDW63316.1 CM000157 EDW88801.1 CH477322 EAT43599.1 CH480818 EDW52410.1 CP012523 ALC39263.1 JXUM01009916 JXUM01009917 JXUM01009918 JXUM01009919 KQ560294 KXJ83220.1 AE014134 BT150148 AAF52919.1 AGR33801.1 CH954177 EDV58516.1 CM000361 CM002910 EDX04512.1 KMY89497.1 GBYB01004458 JAG74225.1 GGMR01015242 MBY27861.1 GGMR01005023 MBY17642.1 UFQT01000745 SSX26899.1 GBHO01019320 GBRD01003419 GDHC01019823 JAG24284.1 JAG62402.1 JAP98805.1 AXCM01000090 ABLF02036488 ATLV01013476 KE524860 KFB38033.1 GGMS01012764 MBY81967.1 AAAB01008944 EAA10322.4 CH475398 EAL29330.2 CH479180 EDW28402.1 CH964276 EDW85342.2 KK107139 EZA57694.1 KQ979608 KYN20470.1 DS235430 EEB15690.1 GL438820 EFN68413.1 KQ978324 KYM95103.1 APCN01000636 ADTU01023735 ADTU01023736 ADTU01023737 ADTU01023738 ADTU01023739 JXJN01003995 QOIP01000003 RLU24644.1 OUUW01000006 SPP81653.1 GBBI01003534 JAC15178.1 SPP81652.1 GGLE01006869 MBY10995.1 CVRI01000055 CRL01191.1 GFJQ02004438 JAW02532.1 GEDV01005911 JAP82646.1 KQ981965 KYN31843.1 GACK01006784 JAA58250.1 KZ288430 PBC25841.1 GAMC01009780 JAB96775.1 CCAG010023386 BT003541 AAO39545.1 KRT05324.1 LJIJ01000793 ODM94540.1
KPI95108.1 NWSH01003361 PCG66478.1 PCG66479.1 PCG66477.1 AGBW02008204 OWR54035.1 KQ461187 KPJ07350.1 KQ971307 EFA10977.1 GEDC01029751 GEDC01018823 JAS07547.1 JAS18475.1 GEZM01079030 JAV62530.1 NEVH01007818 PNF35471.1 APGK01020369 KB740193 KB631602 ENN81148.1 ERL84544.1 PNF35470.1 PNF35472.1 PNF35473.1 AJWK01012548 GEBQ01021816 JAT18161.1 DS232030 EDS32541.1 KQ434879 KZC09954.1 CH933807 EDW13367.1 CH902620 EDV32318.1 GBYB01004457 JAG74224.1 CH916372 EDV98964.1 CH940649 EDW63316.1 CM000157 EDW88801.1 CH477322 EAT43599.1 CH480818 EDW52410.1 CP012523 ALC39263.1 JXUM01009916 JXUM01009917 JXUM01009918 JXUM01009919 KQ560294 KXJ83220.1 AE014134 BT150148 AAF52919.1 AGR33801.1 CH954177 EDV58516.1 CM000361 CM002910 EDX04512.1 KMY89497.1 GBYB01004458 JAG74225.1 GGMR01015242 MBY27861.1 GGMR01005023 MBY17642.1 UFQT01000745 SSX26899.1 GBHO01019320 GBRD01003419 GDHC01019823 JAG24284.1 JAG62402.1 JAP98805.1 AXCM01000090 ABLF02036488 ATLV01013476 KE524860 KFB38033.1 GGMS01012764 MBY81967.1 AAAB01008944 EAA10322.4 CH475398 EAL29330.2 CH479180 EDW28402.1 CH964276 EDW85342.2 KK107139 EZA57694.1 KQ979608 KYN20470.1 DS235430 EEB15690.1 GL438820 EFN68413.1 KQ978324 KYM95103.1 APCN01000636 ADTU01023735 ADTU01023736 ADTU01023737 ADTU01023738 ADTU01023739 JXJN01003995 QOIP01000003 RLU24644.1 OUUW01000006 SPP81653.1 GBBI01003534 JAC15178.1 SPP81652.1 GGLE01006869 MBY10995.1 CVRI01000055 CRL01191.1 GFJQ02004438 JAW02532.1 GEDV01005911 JAP82646.1 KQ981965 KYN31843.1 GACK01006784 JAA58250.1 KZ288430 PBC25841.1 GAMC01009780 JAB96775.1 CCAG010023386 BT003541 AAO39545.1 KRT05324.1 LJIJ01000793 ODM94540.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000007266
+ More
UP000235965 UP000019118 UP000030742 UP000192223 UP000092461 UP000002320 UP000005203 UP000076502 UP000009192 UP000007801 UP000001070 UP000008792 UP000002282 UP000008820 UP000001292 UP000092553 UP000069940 UP000249989 UP000000803 UP000008711 UP000000304 UP000192221 UP000075883 UP000007819 UP000075900 UP000030765 UP000075920 UP000075903 UP000007062 UP000001819 UP000008744 UP000076408 UP000075884 UP000007798 UP000053097 UP000078492 UP000009046 UP000075880 UP000000311 UP000078542 UP000075882 UP000075840 UP000005205 UP000092460 UP000279307 UP000092445 UP000092443 UP000268350 UP000091820 UP000078200 UP000095300 UP000183832 UP000069272 UP000078541 UP000242457 UP000092444 UP000094527
UP000235965 UP000019118 UP000030742 UP000192223 UP000092461 UP000002320 UP000005203 UP000076502 UP000009192 UP000007801 UP000001070 UP000008792 UP000002282 UP000008820 UP000001292 UP000092553 UP000069940 UP000249989 UP000000803 UP000008711 UP000000304 UP000192221 UP000075883 UP000007819 UP000075900 UP000030765 UP000075920 UP000075903 UP000007062 UP000001819 UP000008744 UP000076408 UP000075884 UP000007798 UP000053097 UP000078492 UP000009046 UP000075880 UP000000311 UP000078542 UP000075882 UP000075840 UP000005205 UP000092460 UP000279307 UP000092445 UP000092443 UP000268350 UP000091820 UP000078200 UP000095300 UP000183832 UP000069272 UP000078541 UP000242457 UP000092444 UP000094527
Pfam
Interpro
IPR018980
FERM_PH-like_C
+ More
IPR029071 Ubiquitin-like_domsf
IPR000299 FERM_domain
IPR019747 FERM_CS
IPR018979 FERM_N
IPR011993 PH-like_dom_sf
IPR019748 FERM_central
IPR000798 Ez/rad/moesin-like
IPR014847 FERM-adjacent
IPR035963 FERM_2
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR019749 Band_41_domain
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR036961 Kinesin_motor_dom_sf
IPR001609 Myosin_head_motor_dom
IPR027417 P-loop_NTPase
IPR029071 Ubiquitin-like_domsf
IPR000299 FERM_domain
IPR019747 FERM_CS
IPR018979 FERM_N
IPR011993 PH-like_dom_sf
IPR019748 FERM_central
IPR000798 Ez/rad/moesin-like
IPR014847 FERM-adjacent
IPR035963 FERM_2
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR019749 Band_41_domain
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR036961 Kinesin_motor_dom_sf
IPR001609 Myosin_head_motor_dom
IPR027417 P-loop_NTPase
Gene 3D
ProteinModelPortal
H9J251
A0A2H1VPU7
A0A194PQ68
A0A2A4J485
A0A2A4J3P0
A0A2A4J2V8
+ More
A0A212FJW6 A0A0N0PAM2 D6W6N9 A0A1B6CYS5 A0A1Y1KQS4 A0A2J7R3Q3 N6TSX7 A0A2J7R3S6 A0A2J7R3S0 A0A2J7R3R3 A0A1W4XG86 A0A1W4XRA3 A0A1B0CHP0 A0A1B6L3F3 B0WPV3 A0A087ZS10 A0A154PEU2 B4KHT0 B3MPS0 A0A0C9QUZ1 B4JPY4 B4LVD7 B4NZA6 Q17BE5 B4HWL5 A0A0M3QTP3 A0A182GDD2 Q9VKY7 B3N9M1 B4Q970 A0A0C9PSY0 A0A1W4USX9 A0A2S2PEH3 A0A2S2NL20 A0A336MDB5 A0A0A9XWI4 A0A182MAW3 J9JZI4 A0A182R2J6 A0A084VJ89 A0A2S2QW22 A0A182WA08 A0A182UQW5 Q7Q844 Q29CW6 B4G7K2 A0A182Y9A6 A0A182NT53 B4NLV3 A0A026WRH3 A0A195E5K1 E0VQN4 A0A182IMJ2 E2ADQ1 A0A151I8X7 A0A182LK09 A0A182IB29 A0A158NR13 A0A1B0AV55 A0A3L8DW25 A0A1A9ZM10 A0A1A9Y5P4 A0A3B0JLC9 A0A1A9WJ43 A0A023F1K5 A0A1A9UTA3 A0A3B0JMU0 A0A2R5LNG6 A0A1I8Q1V9 A0A1J1ILS3 A0A1Z5L583 A0A131YWU2 A0A182F2U3 A0A195EUB8 L7M2I0 A0A2A3E434 W8B6G4 A0A1B0FPV8 Q86P10 A0A0R3NWK1 A0A1D2MN79
A0A212FJW6 A0A0N0PAM2 D6W6N9 A0A1B6CYS5 A0A1Y1KQS4 A0A2J7R3Q3 N6TSX7 A0A2J7R3S6 A0A2J7R3S0 A0A2J7R3R3 A0A1W4XG86 A0A1W4XRA3 A0A1B0CHP0 A0A1B6L3F3 B0WPV3 A0A087ZS10 A0A154PEU2 B4KHT0 B3MPS0 A0A0C9QUZ1 B4JPY4 B4LVD7 B4NZA6 Q17BE5 B4HWL5 A0A0M3QTP3 A0A182GDD2 Q9VKY7 B3N9M1 B4Q970 A0A0C9PSY0 A0A1W4USX9 A0A2S2PEH3 A0A2S2NL20 A0A336MDB5 A0A0A9XWI4 A0A182MAW3 J9JZI4 A0A182R2J6 A0A084VJ89 A0A2S2QW22 A0A182WA08 A0A182UQW5 Q7Q844 Q29CW6 B4G7K2 A0A182Y9A6 A0A182NT53 B4NLV3 A0A026WRH3 A0A195E5K1 E0VQN4 A0A182IMJ2 E2ADQ1 A0A151I8X7 A0A182LK09 A0A182IB29 A0A158NR13 A0A1B0AV55 A0A3L8DW25 A0A1A9ZM10 A0A1A9Y5P4 A0A3B0JLC9 A0A1A9WJ43 A0A023F1K5 A0A1A9UTA3 A0A3B0JMU0 A0A2R5LNG6 A0A1I8Q1V9 A0A1J1ILS3 A0A1Z5L583 A0A131YWU2 A0A182F2U3 A0A195EUB8 L7M2I0 A0A2A3E434 W8B6G4 A0A1B0FPV8 Q86P10 A0A0R3NWK1 A0A1D2MN79
PDB
1GG3
E-value=3.76098e-54,
Score=536
Ontologies
GO
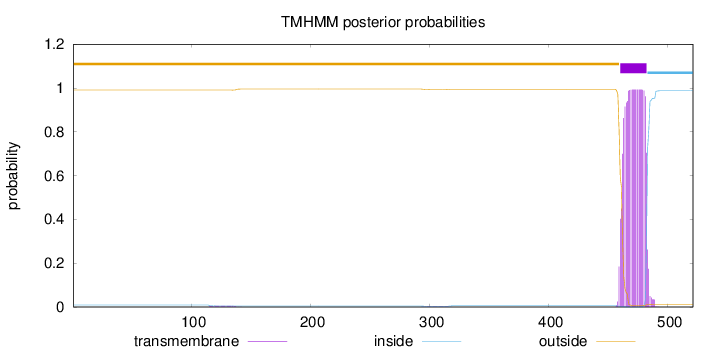
Topology
Length:
521
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.77721
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00945
outside
1 - 459
TMhelix
460 - 482
inside
483 - 521
Population Genetic Test Statistics
Pi
165.414613
Theta
163.429813
Tajima's D
0.986011
CLR
0.411959
CSRT
0.656767161641918
Interpretation
Uncertain
