Gene
KWMTBOMO03005 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003608
Annotation
PREDICTED:_elongation_factor_1-alpha_isoform_X1_[Bombyx_mori]
Full name
Elongation factor 1-alpha
+ More
Elongation factor 1-alpha 1
Elongation factor 1-alpha 1
Alternative Name
50 kDa female-specific protein
Location in the cell
Cytoplasmic Reliability : 4.339
Sequence
CDS
ATGGGCAAGGAAAAGACTCACATTAACATTGTCGTCATCGGACACGTCGACTCCGGCAAGTCCACCACCACTGGTCACTTGATCTACAAATGTGGTGGTATTGACAAACGTACCATCGAGAAGTTCGAGAAGGAGGCCCAGGAAATGGGTAAAGGATCCTTCAAATATGCTTGGGTATTGGACAAACTAAAGGCTGAGCGTGAGCGTGGTATCACAATCGATATTGCTCTCTGGAAGTTCGAAACTAGCAAGTACTATGTTACCATCATTGATGCTCCTGGACACAGAGATTTCATCAAGAACATGATCACAGGAACCTCTCAGGCTGATTGCGCTGTGCTCATCGTAGCTGCCGGTACCGGTGAATTCGAAGCTGGTATCTCTAAGAACGGTCAAACCCGTGAGCATGCCTTGCTCGCTTTCACCCTCGGTGTCAAACAGCTCATCGTAGGAGTAAACAAAATGGATTCCACTGAACCACCATACAGTGAGCCCAGATTTGAGGAAATCAAGAAGGAAGTATCCTCATACATCAAGAAGATTGGCTACAACCCAGCTGCTGTCGCTTTCGTGCCCATTTCTGGATGGCACGGAGACAACATGTTGGAGCCTTCAACCAAAATGCCTTGGTTCAAGGGATGGCAGGTGGAGCGTAAGGAAGGCAAAGCTGACGGAAAATGCCTCATTGAAGCTCTCGATGCCATCCTGCCACCTGCCCGCCCCACTGACAAGCCCCTGCGTCTTCCCCTGCAAGACGTATACAAAATCGGTGGTATTGGTACCGTGCCCGTCGGCAGAGTTGAAACTGGTGTGTTGAAACCAGGTACCATTGTTGTCTTTGCCCCCGCCAACATCACTACTGAAGTCAAGTCTGTGGAGATGCACCACGAAGCTCTCCAAGAAGCTGTACCTGGAGACAATGTAGGTTTCAACGTAAAGAACGTGTCCGTCAAGGAATTGCGTCGTGGTTATGTTGCTGGTGACTCCAAAAACAACCCACCTAAGGGTGCTGCAGATTTTACAGCTCAAGTCATTGTGCTTAACCATCCTGGTCAAATCTCAAACGGTTACACACCAGTCTTGGATTGCCACACTGCCCACATTGCCTGCAAATTTGCAGAAATCAAAGAAAAAGTTGACCGTCGTACTGGTAAATCTACTGAAGTCAACCCAAAATCCATCAAGTCTGGAGATGCAGCCATTGTCAACTTGGTACCTTCCAAGCCTCTATGTGTAGAGTCCTTCCAGGAATTCCCACCCCTCGGTCGTTTTGCTGTCCGTGACATGAGGCAGACAGTTGCTGTCGGAGTCATCAAGGCTGTCAACTTCAAGGAGGCTGGTGGTGGCAAGGTCACTAAAGCTGCCGAAAAGGCCACCAAGGGCAAGAAGTAG
Protein
MGKEKTHINIVVIGHVDSGKSTTTGHLIYKCGGIDKRTIEKFEKEAQEMGKGSFKYAWVLDKLKAERERGITIDIALWKFETSKYYVTIIDAPGHRDFIKNMITGTSQADCAVLIVAAGTGEFEAGISKNGQTREHALLAFTLGVKQLIVGVNKMDSTEPPYSEPRFEEIKKEVSSYIKKIGYNPAAVAFVPISGWHGDNMLEPSTKMPWFKGWQVERKEGKADGKCLIEALDAILPPARPTDKPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGTIVVFAPANITTEVKSVEMHHEALQEAVPGDNVGFNVKNVSVKELRRGYVAGDSKNNPPKGAADFTAQVIVLNHPGQISNGYTPVLDCHTAHIACKFAEIKEKVDRRTGKSTEVNPKSIKSGDAAIVNLVPSKPLCVESFQEFPPLGRFAVRDMRQTVAVGVIKAVNFKEAGGGKVTKAAEKATKGKK
Summary
Description
This protein promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis.
Similarity
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. EF-Tu/EF-1A subfamily.
Keywords
Complete proteome
Cytoplasm
Elongation factor
GTP-binding
Nucleotide-binding
Phosphoprotein
Protein biosynthesis
Reference proteome
Direct protein sequencing
Feature
chain Elongation factor 1-alpha
Uniprot
H9J271
A0A0A0QY84
P29520
A9XXA7
A0A0N1PF32
I4DLT8
+ More
A0A0B5L5V8 A0A194PP74 L7WID6 A0A0K2CTM2 B2DBL0 A0A3B0EBP7 A0A0L7LTH3 A0A385AAY2 A0A2A4JGU8 A0A0Q4U618 S4NNA1 C9E272 A0A385AAX6 A0A385AAX5 A0A385AAX9 A0A385AAY3 A0A385AAY0 A0A385AAZ2 A0A182XYS3 A0A182GCR7 A0A385AAX8 A0A182PQJ0 Q1HR88 A0A182MBB0 A0A385AAY5 C9WPP0 A0A385AAX3 A0A385AAY8 A0A139WNV6 A0A2Z4G644 A0A1Q3F505 A0A212FJY4 Q5QBK9 A0A182R7D5 W5JEK7 A0A182NHZ6 A0A2M3ZYC9 B0WQ61 A0A1Q3F4X7 A0A2M3ZY17 A0A182FHI6 A0A182V4W0 A0A182TV27 A0A182X3V3 A0A182KR72 Q7PT29 A0A182I369 A0A182IMY3 A0A0T6B5E7 A0A182JQ83 A0A2M3ZF36 A0A182WJB9 A0A084WN46 K7IVS1 A0A232ETV4 V5I9P6 A0A0C4URP8 A0A0K8TPH8 Q25BM8 A0A1L8DS46 A0A0J9TZ78 A0A1J1IE44 B3NSB3 A0A182SXI3 P08736 E2A252 A0A1W4VBV4 A0A023ESN1 A0A291L1Q9 A0A0J7KIA4 A0A2H4N0Q7 A0A286T8F0 D6WKQ3 A0A1L8EHJ9 A0A1L8DRV2 B4P5Z5 C9K4M9 B4MDX7 F1SZZ6 A0A1L8EFG4 A0A195CG51 A0A1W4X863 A0A1B0C5C9 B4MYN1 A0A1A9V2G9 B3MG36 A0A069DZ22 A0A151XEX5 A0A336KP24 A0A0C9R131 A0A1Y1L2Z1 A0A0G4AKT1 Q28XB6 A0A288SHG5
A0A0B5L5V8 A0A194PP74 L7WID6 A0A0K2CTM2 B2DBL0 A0A3B0EBP7 A0A0L7LTH3 A0A385AAY2 A0A2A4JGU8 A0A0Q4U618 S4NNA1 C9E272 A0A385AAX6 A0A385AAX5 A0A385AAX9 A0A385AAY3 A0A385AAY0 A0A385AAZ2 A0A182XYS3 A0A182GCR7 A0A385AAX8 A0A182PQJ0 Q1HR88 A0A182MBB0 A0A385AAY5 C9WPP0 A0A385AAX3 A0A385AAY8 A0A139WNV6 A0A2Z4G644 A0A1Q3F505 A0A212FJY4 Q5QBK9 A0A182R7D5 W5JEK7 A0A182NHZ6 A0A2M3ZYC9 B0WQ61 A0A1Q3F4X7 A0A2M3ZY17 A0A182FHI6 A0A182V4W0 A0A182TV27 A0A182X3V3 A0A182KR72 Q7PT29 A0A182I369 A0A182IMY3 A0A0T6B5E7 A0A182JQ83 A0A2M3ZF36 A0A182WJB9 A0A084WN46 K7IVS1 A0A232ETV4 V5I9P6 A0A0C4URP8 A0A0K8TPH8 Q25BM8 A0A1L8DS46 A0A0J9TZ78 A0A1J1IE44 B3NSB3 A0A182SXI3 P08736 E2A252 A0A1W4VBV4 A0A023ESN1 A0A291L1Q9 A0A0J7KIA4 A0A2H4N0Q7 A0A286T8F0 D6WKQ3 A0A1L8EHJ9 A0A1L8DRV2 B4P5Z5 C9K4M9 B4MDX7 F1SZZ6 A0A1L8EFG4 A0A195CG51 A0A1W4X863 A0A1B0C5C9 B4MYN1 A0A1A9V2G9 B3MG36 A0A069DZ22 A0A151XEX5 A0A336KP24 A0A0C9R131 A0A1Y1L2Z1 A0A0G4AKT1 Q28XB6 A0A288SHG5
Pubmed
19121390
8441684
18154650
26354079
22651552
26263512
+ More
18712529 26227816 23622113 25244985 26483478 17204158 17510324 18362917 19820115 29889877 22118469 20920257 23761445 20966253 12364791 14747013 17210077 24438588 20075255 28648823 25692689 26369729 22936249 17994087 16593598 3131735 10731132 12537572 8500545 3135536 20798317 24945155 28964897 17550304 20833722 21167282 23691247 26334808 28004739 26252388 15632085
18712529 26227816 23622113 25244985 26483478 17204158 17510324 18362917 19820115 29889877 22118469 20920257 23761445 20966253 12364791 14747013 17210077 24438588 20075255 28648823 25692689 26369729 22936249 17994087 16593598 3131735 10731132 12537572 8500545 3135536 20798317 24945155 28964897 17550304 20833722 21167282 23691247 26334808 28004739 26252388 15632085
EMBL
BABH01007426
JQ638952
AFJ44727.1
KM027246
AIU94615.1
D13338
+ More
EU016386 ABV68853.1 KQ461187 KPJ07349.1 AK402256 BAM18878.1 KM923784 AJG43843.1 KQ459597 KPI95107.1 KC007373 AGC82213.1 KT218669 ODYU01003719 ALA09389.1 SOQ42848.1 AB264694 BAG30769.1 RBVL01000042 RKO12866.1 JTDY01000116 KOB78760.1 MH592940 AXL66192.1 NWSH01001631 PCG70643.1 LMMH01000027 KQO52249.1 GAIX01012329 JAA80231.1 GQ452009 ACV83782.1 MH592929 AXL66181.1 MH592934 AXL66186.1 MH592937 AXL66189.1 MH592938 AXL66190.1 MH592935 AXL66187.1 MH592942 AXL66194.1 JXUM01054558 KQ561827 KXJ77411.1 MH592933 AXL66185.1 DQ440206 CH477217 ABF18239.1 EJY57378.1 AXCM01017070 MH592941 AXL66193.1 FJ788508 ACU27755.1 MH592928 AXL66180.1 MH592939 AXL66191.1 KQ971307 KYB29556.1 MG983771 AWV96606.1 GFDL01012401 JAV22644.1 AGBW02008204 OWR54036.1 AY752802 UFQT01000004 AAV84215.1 SSX17216.1 ADMH02001732 ETN61259.1 GGFK01000077 MBW33398.1 DS232034 EDS32715.1 GFDL01012424 JAV22621.1 GGFK01000074 MBW33395.1 AAAB01008807 EAA04644.4 APCN01000199 LJIG01009803 KRT82349.1 GGFM01006362 MBW27113.1 ATLV01024562 KE525352 KFB51640.1 NNAY01002218 OXU21782.1 GALX01002570 JAB65896.1 KJ534557 AIY54298.1 GDAI01001306 JAI16297.1 AB253792 BAE91879.1 GFDF01004914 JAV09170.1 CM002911 KMY93055.1 CVRI01000047 CRK98525.1 CH954179 EDV56415.1 M11744 X06869 AE013599 BT011039 BT099573 GL435917 EFN72500.1 GAPW01001320 JAC12278.1 KY809859 ATI09807.1 LBMM01006973 KMQ90153.1 KY653082 ATU47276.1 LC260491 BBA45758.1 KQ971342 EFA03028.1 GFDG01000720 JAV18079.1 GFDF01004915 JAV09169.1 CM000158 EDW90870.1 AB490338 BAH28891.1 CH940662 EDW58742.1 AB591382 BAK08877.2 GFDG01001480 JAV17319.1 KQ977791 KYM99729.1 JXJN01025928 CH963894 EDW77220.1 CH902619 EDV36731.1 GBGD01001395 JAC87494.1 KQ982215 KYQ58929.1 UFQS01000679 UFQT01000679 SSX06137.1 SSX26493.1 GBYB01001685 GBYB01010028 JAG71452.1 JAG79795.1 GEZM01068840 JAV66740.1 KR133396 AKM70866.1 CM000071 EAL26400.2 KX981999 APQ43052.1
EU016386 ABV68853.1 KQ461187 KPJ07349.1 AK402256 BAM18878.1 KM923784 AJG43843.1 KQ459597 KPI95107.1 KC007373 AGC82213.1 KT218669 ODYU01003719 ALA09389.1 SOQ42848.1 AB264694 BAG30769.1 RBVL01000042 RKO12866.1 JTDY01000116 KOB78760.1 MH592940 AXL66192.1 NWSH01001631 PCG70643.1 LMMH01000027 KQO52249.1 GAIX01012329 JAA80231.1 GQ452009 ACV83782.1 MH592929 AXL66181.1 MH592934 AXL66186.1 MH592937 AXL66189.1 MH592938 AXL66190.1 MH592935 AXL66187.1 MH592942 AXL66194.1 JXUM01054558 KQ561827 KXJ77411.1 MH592933 AXL66185.1 DQ440206 CH477217 ABF18239.1 EJY57378.1 AXCM01017070 MH592941 AXL66193.1 FJ788508 ACU27755.1 MH592928 AXL66180.1 MH592939 AXL66191.1 KQ971307 KYB29556.1 MG983771 AWV96606.1 GFDL01012401 JAV22644.1 AGBW02008204 OWR54036.1 AY752802 UFQT01000004 AAV84215.1 SSX17216.1 ADMH02001732 ETN61259.1 GGFK01000077 MBW33398.1 DS232034 EDS32715.1 GFDL01012424 JAV22621.1 GGFK01000074 MBW33395.1 AAAB01008807 EAA04644.4 APCN01000199 LJIG01009803 KRT82349.1 GGFM01006362 MBW27113.1 ATLV01024562 KE525352 KFB51640.1 NNAY01002218 OXU21782.1 GALX01002570 JAB65896.1 KJ534557 AIY54298.1 GDAI01001306 JAI16297.1 AB253792 BAE91879.1 GFDF01004914 JAV09170.1 CM002911 KMY93055.1 CVRI01000047 CRK98525.1 CH954179 EDV56415.1 M11744 X06869 AE013599 BT011039 BT099573 GL435917 EFN72500.1 GAPW01001320 JAC12278.1 KY809859 ATI09807.1 LBMM01006973 KMQ90153.1 KY653082 ATU47276.1 LC260491 BBA45758.1 KQ971342 EFA03028.1 GFDG01000720 JAV18079.1 GFDF01004915 JAV09169.1 CM000158 EDW90870.1 AB490338 BAH28891.1 CH940662 EDW58742.1 AB591382 BAK08877.2 GFDG01001480 JAV17319.1 KQ977791 KYM99729.1 JXJN01025928 CH963894 EDW77220.1 CH902619 EDV36731.1 GBGD01001395 JAC87494.1 KQ982215 KYQ58929.1 UFQS01000679 UFQT01000679 SSX06137.1 SSX26493.1 GBYB01001685 GBYB01010028 JAG71452.1 JAG79795.1 GEZM01068840 JAV66740.1 KR133396 AKM70866.1 CM000071 EAL26400.2 KX981999 APQ43052.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000276569
UP000037510
UP000218220
+ More
UP000051797 UP000076408 UP000069940 UP000249989 UP000075885 UP000008820 UP000075883 UP000007266 UP000007151 UP000075900 UP000000673 UP000075884 UP000002320 UP000069272 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075880 UP000075881 UP000075920 UP000030765 UP000002358 UP000215335 UP000183832 UP000008711 UP000075901 UP000000803 UP000000311 UP000192221 UP000036403 UP000002282 UP000008792 UP000078542 UP000192223 UP000092460 UP000007798 UP000078200 UP000007801 UP000075809 UP000001819
UP000051797 UP000076408 UP000069940 UP000249989 UP000075885 UP000008820 UP000075883 UP000007266 UP000007151 UP000075900 UP000000673 UP000075884 UP000002320 UP000069272 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075880 UP000075881 UP000075920 UP000030765 UP000002358 UP000215335 UP000183832 UP000008711 UP000075901 UP000000803 UP000000311 UP000192221 UP000036403 UP000002282 UP000008792 UP000078542 UP000192223 UP000092460 UP000007798 UP000078200 UP000007801 UP000075809 UP000001819
Pfam
Interpro
IPR004161
EFTu-like_2
+ More
IPR009000 Transl_B-barrel_sf
IPR027417 P-loop_NTPase
IPR031157 G_TR_CS
IPR009001 Transl_elong_EF1A/Init_IF2_C
IPR004160 Transl_elong_EFTu/EF1A_C
IPR000795 TF_GTP-bd_dom
IPR004539 Transl_elong_EF1A_euk/arc
IPR021774 DUF3338
IPR009057 Homeobox-like_sf
IPR001005 SANT/Myb
IPR013219 Ribosomal_S27/S33_mit
IPR013818 Lipase/vitellogenin
IPR029058 AB_hydrolase
IPR009000 Transl_B-barrel_sf
IPR027417 P-loop_NTPase
IPR031157 G_TR_CS
IPR009001 Transl_elong_EF1A/Init_IF2_C
IPR004160 Transl_elong_EFTu/EF1A_C
IPR000795 TF_GTP-bd_dom
IPR004539 Transl_elong_EF1A_euk/arc
IPR021774 DUF3338
IPR009057 Homeobox-like_sf
IPR001005 SANT/Myb
IPR013219 Ribosomal_S27/S33_mit
IPR013818 Lipase/vitellogenin
IPR029058 AB_hydrolase
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J271
A0A0A0QY84
P29520
A9XXA7
A0A0N1PF32
I4DLT8
+ More
A0A0B5L5V8 A0A194PP74 L7WID6 A0A0K2CTM2 B2DBL0 A0A3B0EBP7 A0A0L7LTH3 A0A385AAY2 A0A2A4JGU8 A0A0Q4U618 S4NNA1 C9E272 A0A385AAX6 A0A385AAX5 A0A385AAX9 A0A385AAY3 A0A385AAY0 A0A385AAZ2 A0A182XYS3 A0A182GCR7 A0A385AAX8 A0A182PQJ0 Q1HR88 A0A182MBB0 A0A385AAY5 C9WPP0 A0A385AAX3 A0A385AAY8 A0A139WNV6 A0A2Z4G644 A0A1Q3F505 A0A212FJY4 Q5QBK9 A0A182R7D5 W5JEK7 A0A182NHZ6 A0A2M3ZYC9 B0WQ61 A0A1Q3F4X7 A0A2M3ZY17 A0A182FHI6 A0A182V4W0 A0A182TV27 A0A182X3V3 A0A182KR72 Q7PT29 A0A182I369 A0A182IMY3 A0A0T6B5E7 A0A182JQ83 A0A2M3ZF36 A0A182WJB9 A0A084WN46 K7IVS1 A0A232ETV4 V5I9P6 A0A0C4URP8 A0A0K8TPH8 Q25BM8 A0A1L8DS46 A0A0J9TZ78 A0A1J1IE44 B3NSB3 A0A182SXI3 P08736 E2A252 A0A1W4VBV4 A0A023ESN1 A0A291L1Q9 A0A0J7KIA4 A0A2H4N0Q7 A0A286T8F0 D6WKQ3 A0A1L8EHJ9 A0A1L8DRV2 B4P5Z5 C9K4M9 B4MDX7 F1SZZ6 A0A1L8EFG4 A0A195CG51 A0A1W4X863 A0A1B0C5C9 B4MYN1 A0A1A9V2G9 B3MG36 A0A069DZ22 A0A151XEX5 A0A336KP24 A0A0C9R131 A0A1Y1L2Z1 A0A0G4AKT1 Q28XB6 A0A288SHG5
A0A0B5L5V8 A0A194PP74 L7WID6 A0A0K2CTM2 B2DBL0 A0A3B0EBP7 A0A0L7LTH3 A0A385AAY2 A0A2A4JGU8 A0A0Q4U618 S4NNA1 C9E272 A0A385AAX6 A0A385AAX5 A0A385AAX9 A0A385AAY3 A0A385AAY0 A0A385AAZ2 A0A182XYS3 A0A182GCR7 A0A385AAX8 A0A182PQJ0 Q1HR88 A0A182MBB0 A0A385AAY5 C9WPP0 A0A385AAX3 A0A385AAY8 A0A139WNV6 A0A2Z4G644 A0A1Q3F505 A0A212FJY4 Q5QBK9 A0A182R7D5 W5JEK7 A0A182NHZ6 A0A2M3ZYC9 B0WQ61 A0A1Q3F4X7 A0A2M3ZY17 A0A182FHI6 A0A182V4W0 A0A182TV27 A0A182X3V3 A0A182KR72 Q7PT29 A0A182I369 A0A182IMY3 A0A0T6B5E7 A0A182JQ83 A0A2M3ZF36 A0A182WJB9 A0A084WN46 K7IVS1 A0A232ETV4 V5I9P6 A0A0C4URP8 A0A0K8TPH8 Q25BM8 A0A1L8DS46 A0A0J9TZ78 A0A1J1IE44 B3NSB3 A0A182SXI3 P08736 E2A252 A0A1W4VBV4 A0A023ESN1 A0A291L1Q9 A0A0J7KIA4 A0A2H4N0Q7 A0A286T8F0 D6WKQ3 A0A1L8EHJ9 A0A1L8DRV2 B4P5Z5 C9K4M9 B4MDX7 F1SZZ6 A0A1L8EFG4 A0A195CG51 A0A1W4X863 A0A1B0C5C9 B4MYN1 A0A1A9V2G9 B3MG36 A0A069DZ22 A0A151XEX5 A0A336KP24 A0A0C9R131 A0A1Y1L2Z1 A0A0G4AKT1 Q28XB6 A0A288SHG5
PDB
5LZS
E-value=0,
Score=2032
Ontologies
KEGG
GO
Topology
Subcellular location
Cytoplasm
Secreted
Secreted
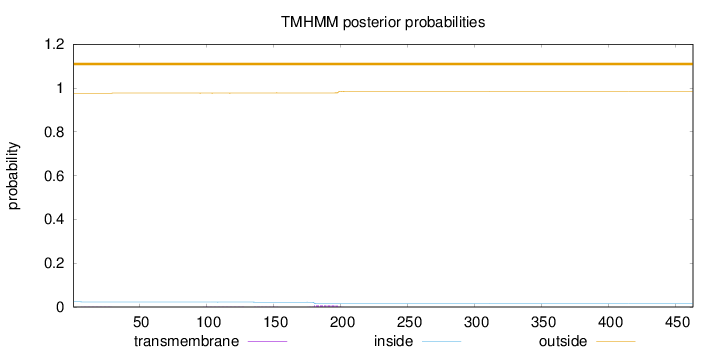
Length:
463
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17776
Exp number, first 60 AAs:
0.02179
Total prob of N-in:
0.02397
outside
1 - 463
Population Genetic Test Statistics
Pi
220.222763
Theta
198.781862
Tajima's D
0.015577
CLR
0.977104
CSRT
0.368381580920954
Interpretation
Uncertain
