Gene
KWMTBOMO03002 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003587
Annotation
protein_disulfide-isomerase_like_protein_ERp57_precursor_[Bombyx_mori]
Full name
Protein disulfide-isomerase
Location in the cell
ER Reliability : 4.636
Sequence
CDS
ATGTTCGGATCACTAAAGTTTGTTCTTTTATTAGGCATAATTTATTTATGTAAAGCGGCCGAGGAGGATGTTCTCGATCTTACAGATTCCGACTTTTCGGCTGTTTTATCTCAACATGATACAGCCCTGGTCATGTTTTACGCGCCTTGGTGCGGTCATTGTAAGAGACTAAAGCCAGAGTATGCGGTAGCCGCCGGCCTGTTAAAGACCGATGTCCCTCCGGTGGCGCTAGCTAAAGTAGACTGTACGGAAGGTGGCAAGAGTACTTGTGAACAATTCTCTGTGTCCGGATATCCTACACTGAAAATATTCAGAAAAGGAGAGCTTTCTTCCGAGTACAATGGACCAAGGGAGTCTAATGGCATTGTCAAGTACATGCGTGCCCAAGTTGGACCCAGCTCCAAGGAGCTCCTTACAGTAGCTGATTTTGAAGCTTTTACTTCTAAGGATGAAGTAGTGGTTGTCGGATTCTTCGAAAAGGAATCTGACCTGAAAGGAGAGTTCCTGAAAACAGCTGACAAATTAAGGGAGGAGGTTACTTTTGCTCATTCATCAGCCAATGAAGTTCTTGAAAAAACCGGATACAAAAACAATGTGGTCTTGTATCGTCCCAAGCGACTCCAGAACAAGTTTGAAGACTCATCTGTTGCCTTCGACGGAGACACCGAGAAAGTGTCACTCAAAGCTTTCATCAAGGAGAACTATCACGGTCTGGTCGGCGTTCGCCAAAAGGACAACATCCATGATTTCAGCAACCCCCTTATTGTCGCTTACTATGATGTTGATTATACCAAGAACCCGAAGGGCACTAACTACTGGAGGAACCGTGTCCTCAAGGTAGCCAAGGAGCAAACCGAGGCGACGTTCGCAGTCAGCGACAAGGACGACTTCACGCACGAGCTCAACGAGTTCGGCATCGACTTCGCTAAAGGCGACAAGCCCGTAGTGGCCGGCCGCGACGCAGACGGAAACAAATTCGTCATGAGCGCGGAGTTCAGCATCGAAAATCTACTAACGTTCACAAAGGACCTGCTCGATGGCAAGCTGGAGCCATTCGTGAAATCCGAGGCCATACCGGAAAACGACGGCCCAGTGAAGGTAGCCGTCGGCAAGAACTTCAAGGAACTGGTCACGGACAGCAACAGGGACGCACTCATAGAATTCTATGCGCCATGGTGCGGACACTGTCAGAAGCTTGCTCCCGTCTGGGAAGAGCTTGGTGAAAAGTTGAAAGATGAAGAAGTTGACATCATTAAGATCGATGCAACTGCAAATGACTGGCCCAAATCACAATTCGACGTTTCCGGCTTCCCCACAATCTTCTGGAAGCCTAAGGACAGCTCCAAGAAACCTCAGAGATATAATGGAGGACGTGCTCTAGAGGACTTCATCAAGTATGTTTCTGAACAAGCCACAAGCGAACTTAAGGGCTGGGACAGAAAGGGTAACGCCAAACAAGGAAAGGAAGAGTTGTAG
Protein
MFGSLKFVLLLGIIYLCKAAEEDVLDLTDSDFSAVLSQHDTALVMFYAPWCGHCKRLKPEYAVAAGLLKTDVPPVALAKVDCTEGGKSTCEQFSVSGYPTLKIFRKGELSSEYNGPRESNGIVKYMRAQVGPSSKELLTVADFEAFTSKDEVVVVGFFEKESDLKGEFLKTADKLREEVTFAHSSANEVLEKTGYKNNVVLYRPKRLQNKFEDSSVAFDGDTEKVSLKAFIKENYHGLVGVRQKDNIHDFSNPLIVAYYDVDYTKNPKGTNYWRNRVLKVAKEQTEATFAVSDKDDFTHELNEFGIDFAKGDKPVVAGRDADGNKFVMSAEFSIENLLTFTKDLLDGKLEPFVKSEAIPENDGPVKVAVGKNFKELVTDSNRDALIEFYAPWCGHCQKLAPVWEELGEKLKDEEVDIIKIDATANDWPKSQFDVSGFPTIFWKPKDSSKKPQRYNGGRALEDFIKYVSEQATSELKGWDRKGNAKQGKEEL
Summary
Catalytic Activity
Catalyzes the rearrangement of -S-S- bonds in proteins.
Similarity
Belongs to the protein disulfide isomerase family.
Feature
chain Protein disulfide-isomerase
Uniprot
Q587N3
Q587N4
A0A2A4JF99
A0A0N1PHN3
H9J250
A0A194PVF1
+ More
A0A212FGT6 I4DIY4 S4PKX7 A0A2J7R3Z5 A0A1V0QBC3 A0A0K8TPP3 D6W727 G9C5C9 E0VQN7 A0A1B6CLZ3 A0A1Y1K4Q8 A0A1B6EMG6 Q17L92 A0A1B6HL77 A0A1W4WNQ6 A0A1J1IH13 A0A1S4EYS5 A0A2M3YY74 A0A0P4WFJ6 B2ZPN7 A0A2M3ZZB8 A0A1L8DTR3 A0A2M3ZED9 A0A1L8DTX7 A0A023EVX9 A0A2M3ZZB6 A0A182H2M9 A0A1I9WL43 A0A023EU67 A0A182LV82 A0A182FHJ0 A0A336KRW2 T1E811 A0A2M3ZZK8 T1DI34 T1PHC1 A0A182R7C8 W5JD33 Q3YMT9 A0A1W4VCW7 A0A023ET60 A0A1I8N351 A0A182XYR3 B4QC45 B3NSB4 A0A182WJC8 A0A084WN41 A0A182PQK0 Q3YMU0 A0A182V7X7 A0A182JR00 E9IY48 B4P5Z6 Q9TWZ1 B3MG35 B4HNU6 J3JWJ9 A0A1Q3G208 K7J8B8 A0A1Y9H9J2 A0A0J7NV70 A0A0M3QUI7 A0A240PLG7 A0A182TUT7 N6U0U4 A0A026WCE2 U4U691 A0A182NI02 A0A182X3W2 Q5TWW9 A0A182I360 J9JYS8 R4WP97 A0A182UBC7 A0A195CZV3 A0A154PLS6 A0A0P5Q3I4 A0A034WXY3 A0A0P6E5T3 A0A195BRK6 A0A0T6BBT9 A0A0P5L3J3 A0A151X3E7 A0A2H8TTB8 A0A3B0JHX0 B4MYN2 A0A0P6FVC3 A0A158NWU6 A0A0A1XSS1 A0A0P5MWN5 A0A1I8NRA8 A0A0K8U8G4 F4WQS3
A0A212FGT6 I4DIY4 S4PKX7 A0A2J7R3Z5 A0A1V0QBC3 A0A0K8TPP3 D6W727 G9C5C9 E0VQN7 A0A1B6CLZ3 A0A1Y1K4Q8 A0A1B6EMG6 Q17L92 A0A1B6HL77 A0A1W4WNQ6 A0A1J1IH13 A0A1S4EYS5 A0A2M3YY74 A0A0P4WFJ6 B2ZPN7 A0A2M3ZZB8 A0A1L8DTR3 A0A2M3ZED9 A0A1L8DTX7 A0A023EVX9 A0A2M3ZZB6 A0A182H2M9 A0A1I9WL43 A0A023EU67 A0A182LV82 A0A182FHJ0 A0A336KRW2 T1E811 A0A2M3ZZK8 T1DI34 T1PHC1 A0A182R7C8 W5JD33 Q3YMT9 A0A1W4VCW7 A0A023ET60 A0A1I8N351 A0A182XYR3 B4QC45 B3NSB4 A0A182WJC8 A0A084WN41 A0A182PQK0 Q3YMU0 A0A182V7X7 A0A182JR00 E9IY48 B4P5Z6 Q9TWZ1 B3MG35 B4HNU6 J3JWJ9 A0A1Q3G208 K7J8B8 A0A1Y9H9J2 A0A0J7NV70 A0A0M3QUI7 A0A240PLG7 A0A182TUT7 N6U0U4 A0A026WCE2 U4U691 A0A182NI02 A0A182X3W2 Q5TWW9 A0A182I360 J9JYS8 R4WP97 A0A182UBC7 A0A195CZV3 A0A154PLS6 A0A0P5Q3I4 A0A034WXY3 A0A0P6E5T3 A0A195BRK6 A0A0T6BBT9 A0A0P5L3J3 A0A151X3E7 A0A2H8TTB8 A0A3B0JHX0 B4MYN2 A0A0P6FVC3 A0A158NWU6 A0A0A1XSS1 A0A0P5MWN5 A0A1I8NRA8 A0A0K8U8G4 F4WQS3
EC Number
5.3.4.1
Pubmed
26354079
19121390
22118469
22651552
23622113
28356351
+ More
26369729 18362917 19820115 21835214 20566863 28004739 17510324 24945155 26483478 27538518 24330624 20920257 23761445 15917496 25315136 25244985 17994087 22936249 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 17550304 8687406 22516182 20075255 23537049 24508170 30249741 12364791 14747013 17210077 23691247 25348373 21347285 25830018 21719571
26369729 18362917 19820115 21835214 20566863 28004739 17510324 24945155 26483478 27538518 24330624 20920257 23761445 15917496 25315136 25244985 17994087 22936249 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 17550304 8687406 22516182 20075255 23537049 24508170 30249741 12364791 14747013 17210077 23691247 25348373 21347285 25830018 21719571
EMBL
AB210112
BAD93614.1
AB210111
BAD93613.1
NWSH01001631
PCG70641.1
+ More
KQ461187 KPJ07347.1 BABH01007426 KQ459597 KPI95105.1 AGBW02008594 OWR52961.1 AK401252 BAM17874.1 GAIX01004300 JAA88260.1 NEVH01007818 PNF35559.1 KX683312 ARE59259.1 GDAI01001505 JAI16098.1 KQ971307 EFA11421.1 HQ851370 AEV89748.1 DS235430 EEB15693.1 GEDC01022987 JAS14311.1 GEZM01095185 JAV55471.1 GECZ01030664 GECZ01024813 GECZ01022011 GECZ01014873 GECZ01012784 JAS39105.1 JAS44956.1 JAS47758.1 JAS54896.1 JAS56985.1 CH477217 EAT47483.1 GECU01032273 JAS75433.1 CVRI01000051 CRK99496.1 GGFM01000472 MBW21223.1 GDRN01053050 JAI66204.1 EU679503 ACD44938.1 GGFK01000520 MBW33841.1 GFDF01004238 JAV09846.1 GGFM01006049 MBW26800.1 GFDF01004237 JAV09847.1 GAPW01001124 JAC12474.1 GGFK01000524 MBW33845.1 JXUM01023842 KQ560673 KXJ81333.1 KU932227 MF278688 APA33863.1 ASL04994.1 GAPW01001126 JAC12472.1 AXCM01001984 UFQS01000899 UFQT01000899 SSX07566.1 SSX27906.1 GAMD01003158 JAA98432.1 GGFK01000591 MBW33912.1 GALA01001176 JAA93676.1 KA647510 AFP62139.1 ADMH02001732 ETN61268.1 DQ062787 AAY56660.1 GAPW01001125 JAC12473.1 CM000362 CM002911 EDX06676.1 KMY93054.1 CH954179 EDV56416.1 ATLV01024562 KE525352 KFB51635.1 AE013599 DQ062786 BT099537 AAF58609.2 AAY56659.1 ACU32621.1 GL766908 EFZ14477.1 CM000158 EDW90871.1 CH902619 EDV36730.1 CH480816 EDW47461.1 BT127617 AEE62579.1 GFDL01001177 JAV33868.1 AAZX01014127 AXCN02000386 LBMM01001449 KMQ96290.1 CP012524 ALC40739.1 APGK01047206 KB741077 ENN74226.1 KK107274 QOIP01000008 EZA53740.1 RLU19705.1 KB631792 ERL86146.1 AAAB01008807 EAL41801.2 APCN01000199 ABLF02031932 AK417486 BAN20701.1 KQ977041 KYN06137.1 KQ434977 KZC12816.1 GDIQ01140688 JAL11038.1 GAKP01000304 JAC58648.1 GDIQ01068613 GDIQ01045929 LRGB01001764 JAN26124.1 KZS10564.1 KQ976424 KYM88584.1 LJIG01002144 KRT84796.1 GDIQ01176906 JAK74819.1 KQ982562 KYQ54933.1 GFXV01005642 MBW17447.1 OUUW01000001 SPP74980.1 CH963894 EDW77221.1 GDIQ01043869 JAN50868.1 ADTU01028568 GBXI01000352 JAD13940.1 GDIQ01154428 JAK97297.1 GDHF01029674 GDHF01023347 GDHF01002536 JAI22640.1 JAI28967.1 JAI49778.1 GL888275 EGI63436.1
KQ461187 KPJ07347.1 BABH01007426 KQ459597 KPI95105.1 AGBW02008594 OWR52961.1 AK401252 BAM17874.1 GAIX01004300 JAA88260.1 NEVH01007818 PNF35559.1 KX683312 ARE59259.1 GDAI01001505 JAI16098.1 KQ971307 EFA11421.1 HQ851370 AEV89748.1 DS235430 EEB15693.1 GEDC01022987 JAS14311.1 GEZM01095185 JAV55471.1 GECZ01030664 GECZ01024813 GECZ01022011 GECZ01014873 GECZ01012784 JAS39105.1 JAS44956.1 JAS47758.1 JAS54896.1 JAS56985.1 CH477217 EAT47483.1 GECU01032273 JAS75433.1 CVRI01000051 CRK99496.1 GGFM01000472 MBW21223.1 GDRN01053050 JAI66204.1 EU679503 ACD44938.1 GGFK01000520 MBW33841.1 GFDF01004238 JAV09846.1 GGFM01006049 MBW26800.1 GFDF01004237 JAV09847.1 GAPW01001124 JAC12474.1 GGFK01000524 MBW33845.1 JXUM01023842 KQ560673 KXJ81333.1 KU932227 MF278688 APA33863.1 ASL04994.1 GAPW01001126 JAC12472.1 AXCM01001984 UFQS01000899 UFQT01000899 SSX07566.1 SSX27906.1 GAMD01003158 JAA98432.1 GGFK01000591 MBW33912.1 GALA01001176 JAA93676.1 KA647510 AFP62139.1 ADMH02001732 ETN61268.1 DQ062787 AAY56660.1 GAPW01001125 JAC12473.1 CM000362 CM002911 EDX06676.1 KMY93054.1 CH954179 EDV56416.1 ATLV01024562 KE525352 KFB51635.1 AE013599 DQ062786 BT099537 AAF58609.2 AAY56659.1 ACU32621.1 GL766908 EFZ14477.1 CM000158 EDW90871.1 CH902619 EDV36730.1 CH480816 EDW47461.1 BT127617 AEE62579.1 GFDL01001177 JAV33868.1 AAZX01014127 AXCN02000386 LBMM01001449 KMQ96290.1 CP012524 ALC40739.1 APGK01047206 KB741077 ENN74226.1 KK107274 QOIP01000008 EZA53740.1 RLU19705.1 KB631792 ERL86146.1 AAAB01008807 EAL41801.2 APCN01000199 ABLF02031932 AK417486 BAN20701.1 KQ977041 KYN06137.1 KQ434977 KZC12816.1 GDIQ01140688 JAL11038.1 GAKP01000304 JAC58648.1 GDIQ01068613 GDIQ01045929 LRGB01001764 JAN26124.1 KZS10564.1 KQ976424 KYM88584.1 LJIG01002144 KRT84796.1 GDIQ01176906 JAK74819.1 KQ982562 KYQ54933.1 GFXV01005642 MBW17447.1 OUUW01000001 SPP74980.1 CH963894 EDW77221.1 GDIQ01043869 JAN50868.1 ADTU01028568 GBXI01000352 JAD13940.1 GDIQ01154428 JAK97297.1 GDHF01029674 GDHF01023347 GDHF01002536 JAI22640.1 JAI28967.1 JAI49778.1 GL888275 EGI63436.1
Proteomes
UP000218220
UP000053240
UP000005204
UP000053268
UP000007151
UP000235965
+ More
UP000007266 UP000009046 UP000008820 UP000192223 UP000183832 UP000069940 UP000249989 UP000075883 UP000069272 UP000075900 UP000000673 UP000192221 UP000095301 UP000076408 UP000000304 UP000008711 UP000075920 UP000030765 UP000075885 UP000000803 UP000075903 UP000075881 UP000002282 UP000007801 UP000001292 UP000002358 UP000075886 UP000036403 UP000092553 UP000075880 UP000075902 UP000019118 UP000053097 UP000279307 UP000030742 UP000075884 UP000076407 UP000007062 UP000075840 UP000007819 UP000078542 UP000076502 UP000076858 UP000078540 UP000075809 UP000268350 UP000007798 UP000005205 UP000095300 UP000007755
UP000007266 UP000009046 UP000008820 UP000192223 UP000183832 UP000069940 UP000249989 UP000075883 UP000069272 UP000075900 UP000000673 UP000192221 UP000095301 UP000076408 UP000000304 UP000008711 UP000075920 UP000030765 UP000075885 UP000000803 UP000075903 UP000075881 UP000002282 UP000007801 UP000001292 UP000002358 UP000075886 UP000036403 UP000092553 UP000075880 UP000075902 UP000019118 UP000053097 UP000279307 UP000030742 UP000075884 UP000076407 UP000007062 UP000075840 UP000007819 UP000078542 UP000076502 UP000076858 UP000078540 UP000075809 UP000268350 UP000007798 UP000005205 UP000095300 UP000007755
Pfam
PF00085 Thioredoxin
Interpro
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
Q587N3
Q587N4
A0A2A4JF99
A0A0N1PHN3
H9J250
A0A194PVF1
+ More
A0A212FGT6 I4DIY4 S4PKX7 A0A2J7R3Z5 A0A1V0QBC3 A0A0K8TPP3 D6W727 G9C5C9 E0VQN7 A0A1B6CLZ3 A0A1Y1K4Q8 A0A1B6EMG6 Q17L92 A0A1B6HL77 A0A1W4WNQ6 A0A1J1IH13 A0A1S4EYS5 A0A2M3YY74 A0A0P4WFJ6 B2ZPN7 A0A2M3ZZB8 A0A1L8DTR3 A0A2M3ZED9 A0A1L8DTX7 A0A023EVX9 A0A2M3ZZB6 A0A182H2M9 A0A1I9WL43 A0A023EU67 A0A182LV82 A0A182FHJ0 A0A336KRW2 T1E811 A0A2M3ZZK8 T1DI34 T1PHC1 A0A182R7C8 W5JD33 Q3YMT9 A0A1W4VCW7 A0A023ET60 A0A1I8N351 A0A182XYR3 B4QC45 B3NSB4 A0A182WJC8 A0A084WN41 A0A182PQK0 Q3YMU0 A0A182V7X7 A0A182JR00 E9IY48 B4P5Z6 Q9TWZ1 B3MG35 B4HNU6 J3JWJ9 A0A1Q3G208 K7J8B8 A0A1Y9H9J2 A0A0J7NV70 A0A0M3QUI7 A0A240PLG7 A0A182TUT7 N6U0U4 A0A026WCE2 U4U691 A0A182NI02 A0A182X3W2 Q5TWW9 A0A182I360 J9JYS8 R4WP97 A0A182UBC7 A0A195CZV3 A0A154PLS6 A0A0P5Q3I4 A0A034WXY3 A0A0P6E5T3 A0A195BRK6 A0A0T6BBT9 A0A0P5L3J3 A0A151X3E7 A0A2H8TTB8 A0A3B0JHX0 B4MYN2 A0A0P6FVC3 A0A158NWU6 A0A0A1XSS1 A0A0P5MWN5 A0A1I8NRA8 A0A0K8U8G4 F4WQS3
A0A212FGT6 I4DIY4 S4PKX7 A0A2J7R3Z5 A0A1V0QBC3 A0A0K8TPP3 D6W727 G9C5C9 E0VQN7 A0A1B6CLZ3 A0A1Y1K4Q8 A0A1B6EMG6 Q17L92 A0A1B6HL77 A0A1W4WNQ6 A0A1J1IH13 A0A1S4EYS5 A0A2M3YY74 A0A0P4WFJ6 B2ZPN7 A0A2M3ZZB8 A0A1L8DTR3 A0A2M3ZED9 A0A1L8DTX7 A0A023EVX9 A0A2M3ZZB6 A0A182H2M9 A0A1I9WL43 A0A023EU67 A0A182LV82 A0A182FHJ0 A0A336KRW2 T1E811 A0A2M3ZZK8 T1DI34 T1PHC1 A0A182R7C8 W5JD33 Q3YMT9 A0A1W4VCW7 A0A023ET60 A0A1I8N351 A0A182XYR3 B4QC45 B3NSB4 A0A182WJC8 A0A084WN41 A0A182PQK0 Q3YMU0 A0A182V7X7 A0A182JR00 E9IY48 B4P5Z6 Q9TWZ1 B3MG35 B4HNU6 J3JWJ9 A0A1Q3G208 K7J8B8 A0A1Y9H9J2 A0A0J7NV70 A0A0M3QUI7 A0A240PLG7 A0A182TUT7 N6U0U4 A0A026WCE2 U4U691 A0A182NI02 A0A182X3W2 Q5TWW9 A0A182I360 J9JYS8 R4WP97 A0A182UBC7 A0A195CZV3 A0A154PLS6 A0A0P5Q3I4 A0A034WXY3 A0A0P6E5T3 A0A195BRK6 A0A0T6BBT9 A0A0P5L3J3 A0A151X3E7 A0A2H8TTB8 A0A3B0JHX0 B4MYN2 A0A0P6FVC3 A0A158NWU6 A0A0A1XSS1 A0A0P5MWN5 A0A1I8NRA8 A0A0K8U8G4 F4WQS3
PDB
6ENY
E-value=2.69245e-114,
Score=1055
Ontologies
GO
Topology
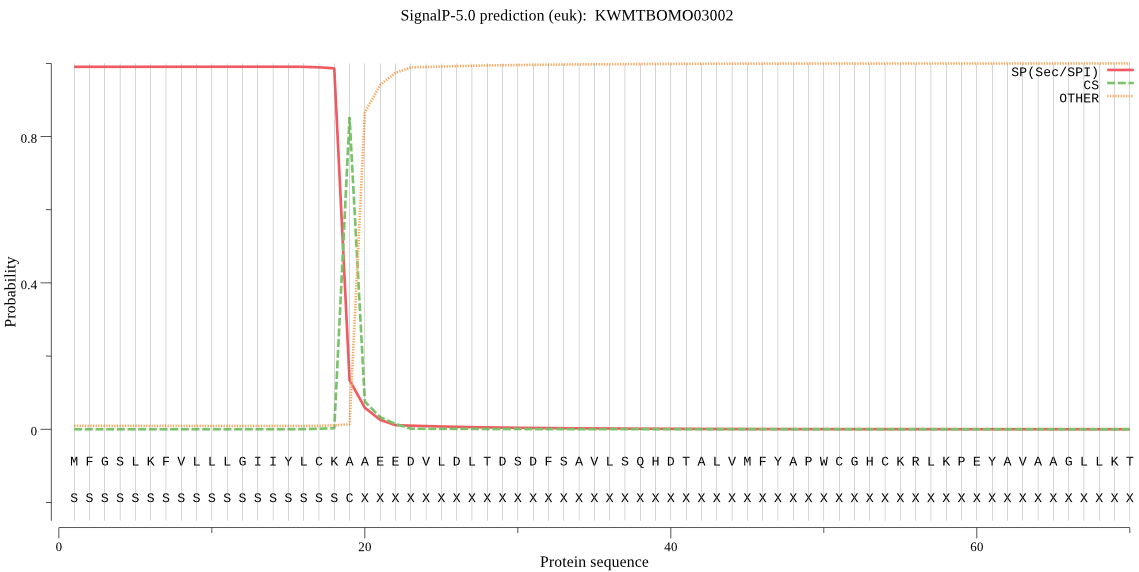
SignalP
Position: 1 - 19,
Likelihood: 0.990476
Length:
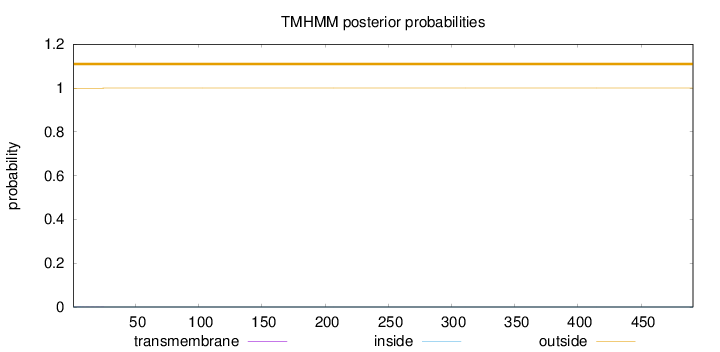
491
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00827999999999999
Exp number, first 60 AAs:
0.00827999999999999
Total prob of N-in:
0.00060
outside
1 - 491
Population Genetic Test Statistics
Pi
238.91798
Theta
167.885338
Tajima's D
1.190783
CLR
0.2673
CSRT
0.714464276786161
Interpretation
Uncertain
