Gene
KWMTBOMO03001 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003610
Annotation
PREDICTED:_glucose-6-phosphate_1-epimerase_[Bombyx_mori]
Full name
Glucose-6-phosphate 1-epimerase
Location in the cell
Cytoplasmic Reliability : 1.439
Sequence
CDS
ATGGCAGCAACGAGTGTTGTCGTTCTCGACAGAGGCAACAATACAACATGTACCGTAAATCTTTTCGGTGCTACTGTAGTGTCATGGAGGGTGAACAACCAAGAGCAATTATTTGTTAGCAAACAAGCCGTTTTCGACGGCAAGAGGGCAATACGAGGCGGTATACCATTCGTTTTTCCTCAATTCGGGCAATGGGCCTTCGGACCTCAGCACGGCTTCGCGCGGGTGGCCCGCTGGCACGTCGAGAAGATGCCCGAGCGACTGCCCAGCGGCGACGTGGAAGCTGTCTTCAGCCTTATGGACGATGAGTTCACCAGGTCGATGTGGCACTTTCAGTTTCGGCTGACGTACCGGCTGATCTTGCGTGAGAAGGAGCTCCACTTCAACATCGGCGTGTACAACCCGAGCAAGGAGCTGACGTTTAGCTGTCAGTTGCTGCTCCACACTTACTTCAAGGTGCCGGACGTGCGCCGCTGTCAGATCACCGGCATGCACGGCTGCATGTTCATTGACAAGACGCGGGAAGGTGCCGTGTACCAGGAGACCCGCGAGGTGGTCACGGTCGGCGAGTGGACGGACCGCATCTACCAGAACACGATGCAGGAGCACATCATAACGAACGTTGTCAGCGGGCGGAAGATGCGGATACAGAAGTACAACTTCCCCGATACAGTGATCTGGAACCCGTGGTCGGAGTACGCGAAGGAGATCCCGGACTTCGGTGACGACGAGTTCCCCAACATGGTGTGCGTGGAGGCCGGGCGGGTGGCCGCGCCCATCGTGTTGCTCCCCGGCACCGCGTTCGAAGCCAGCCAGATCCTCCAGGTAATGTAG
Protein
MAATSVVVLDRGNNTTCTVNLFGATVVSWRVNNQEQLFVSKQAVFDGKRAIRGGIPFVFPQFGQWAFGPQHGFARVARWHVEKMPERLPSGDVEAVFSLMDDEFTRSMWHFQFRLTYRLILREKELHFNIGVYNPSKELTFSCQLLLHTYFKVPDVRRCQITGMHGCMFIDKTREGAVYQETREVVTVGEWTDRIYQNTMQEHIITNVVSGRKMRIQKYNFPDTVIWNPWSEYAKEIPDFGDDEFPNMVCVEAGRVAAPIVLLPGTAFEASQILQVM
Summary
Similarity
Belongs to the glucose-6-phosphate 1-epimerase family.
Uniprot
A0A2A4JGU4
A0A212FGV0
A0A194PQT3
A0A0N1I6F9
E0VQN5
A0A1D2NEP7
+ More
A0A1Y1N837 A0A1B6KHY0 N6T9D7 A0A0T6AXF4 A0A087ZS11 A0A1L8E1N7 K7J8C0 A0A151I8W3 A0A1B6CES6 A0A0C9R6S1 A0A2H1W608 A0A154PDI4 A0A336MCB6 A0A182JN94 A0A1Q3FU28 A0A2M4CVM5 A0A2M4AF02 B4MYQ7 A0A226EZP0 A0A182RWF3 A0A2M4CVN3 E9GXW6 A0A1J1IR13 A0A310SGX2 A0A0M5IZA1 A0A0A1XF52 A0A1S4GZX6 A0A158NR61 Q29K64 B4GWY5 A0A0A1WLB8 A0A3B0KJY9 W8BQK5 A0A034WAT8 A0A0K8UDW8 W8BJ73 D6W7E6 A0A0P6HK98 A0A0R3P0T1 A0A0L0C762 A0A1A9WU95 A0A1I8P4I9 A0A1I8NBW9 B4P3P3 Q9V3D1 B4HX88 B4Q552 B3NAE4 Q8IP67 B3MV55 A0A1W4V7F1 A0A0P4VZ16 R4G5G2 A0A0J9R1L5 X2JAD3 A0A0K8S6M4 A0A0R1DKC4 A0A0Q5WM37 A0A1W4UV42 A0A1W4V8S6 A0A0P8Y5D5 B4M8K3 B4KIW8 A0A1B6GSJ5 A0A0Q9W7C9 A0A0P5AHW8 A0A0Q9XJF1 A0A0P4WIR6 A0A1B0A4J8 A0A3L8DYA6 H9J273 A0A1B0FFA6 A0A0N8E7J3 A0A1I8NBY4 A0A067QUV3 A0A1B0C763 E2ADQ2 A0A0P4X4F1 A0A1B0CQ74 A0A2A3E2B9 A0A023EKN2 A0A2M3ZCU3 A0A026WRG9 A0A232EY56 A0A2P8Y235 Q1DH24 A0A182VNL7 A0A2C9GRI6 Q172P9 A0A182VSJ6 A0A182X882 A0A182P2W2 Q7PW99 A0A182QZQ3
A0A1Y1N837 A0A1B6KHY0 N6T9D7 A0A0T6AXF4 A0A087ZS11 A0A1L8E1N7 K7J8C0 A0A151I8W3 A0A1B6CES6 A0A0C9R6S1 A0A2H1W608 A0A154PDI4 A0A336MCB6 A0A182JN94 A0A1Q3FU28 A0A2M4CVM5 A0A2M4AF02 B4MYQ7 A0A226EZP0 A0A182RWF3 A0A2M4CVN3 E9GXW6 A0A1J1IR13 A0A310SGX2 A0A0M5IZA1 A0A0A1XF52 A0A1S4GZX6 A0A158NR61 Q29K64 B4GWY5 A0A0A1WLB8 A0A3B0KJY9 W8BQK5 A0A034WAT8 A0A0K8UDW8 W8BJ73 D6W7E6 A0A0P6HK98 A0A0R3P0T1 A0A0L0C762 A0A1A9WU95 A0A1I8P4I9 A0A1I8NBW9 B4P3P3 Q9V3D1 B4HX88 B4Q552 B3NAE4 Q8IP67 B3MV55 A0A1W4V7F1 A0A0P4VZ16 R4G5G2 A0A0J9R1L5 X2JAD3 A0A0K8S6M4 A0A0R1DKC4 A0A0Q5WM37 A0A1W4UV42 A0A1W4V8S6 A0A0P8Y5D5 B4M8K3 B4KIW8 A0A1B6GSJ5 A0A0Q9W7C9 A0A0P5AHW8 A0A0Q9XJF1 A0A0P4WIR6 A0A1B0A4J8 A0A3L8DYA6 H9J273 A0A1B0FFA6 A0A0N8E7J3 A0A1I8NBY4 A0A067QUV3 A0A1B0C763 E2ADQ2 A0A0P4X4F1 A0A1B0CQ74 A0A2A3E2B9 A0A023EKN2 A0A2M3ZCU3 A0A026WRG9 A0A232EY56 A0A2P8Y235 Q1DH24 A0A182VNL7 A0A2C9GRI6 Q172P9 A0A182VSJ6 A0A182X882 A0A182P2W2 Q7PW99 A0A182QZQ3
EC Number
5.1.3.15
Pubmed
22118469
26354079
20566863
27289101
28004739
23537049
+ More
20075255 17994087 21292972 25830018 12364791 21347285 15632085 23185243 24495485 25348373 18362917 19820115 26108605 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 27129103 26823975 18057021 30249741 19121390 24845553 20798317 24945155 24508170 28648823 29403074 17510324
20075255 17994087 21292972 25830018 12364791 21347285 15632085 23185243 24495485 25348373 18362917 19820115 26108605 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 27129103 26823975 18057021 30249741 19121390 24845553 20798317 24945155 24508170 28648823 29403074 17510324
EMBL
NWSH01001631
PCG70640.1
AGBW02008594
OWR52962.1
KQ459597
KPI95104.1
+ More
KQ461187 KPJ07346.1 DS235430 EEB15691.1 LJIJ01000065 ODN03740.1 GEZM01011141 GEZM01011140 JAV93638.1 GEBQ01028915 JAT11062.1 APGK01039146 KB740967 KB632399 ENN76864.1 ERL94648.1 LJIG01022567 KRT79888.1 GFDF01001463 JAV12621.1 AAZX01003471 AAZX01016222 AAZX01018366 KQ978324 KYM95101.1 GEDC01025354 JAS11944.1 GBYB01008499 GBYB01008502 JAG78266.1 JAG78269.1 ODYU01006498 SOQ48386.1 KQ434879 KZC09966.1 UFQS01000752 UFQT01000752 SSX06634.1 SSX26981.1 GFDL01003980 JAV31065.1 GGFL01005214 MBW69392.1 GGFK01006048 MBW39369.1 CH963913 EDW77246.1 LNIX01000001 OXA62600.1 GGFL01005215 MBW69393.1 GL732573 EFX75730.1 CVRI01000058 CRL02671.1 KQ760118 OAD61917.1 CP012523 ALC40198.1 GBXI01011969 GBXI01004731 JAD02323.1 JAD09561.1 AAAB01008984 ADTU01023732 ADTU01023733 ADTU01023734 ADTU01023735 CH379061 EAL33314.1 KRT04895.1 CH479195 EDW27312.1 GBXI01014488 JAC99803.1 OUUW01000010 SPP85461.1 GAMC01005214 GAMC01005213 JAC01343.1 GAKP01006278 GAKP01006277 GAKP01006276 JAC52676.1 GDHF01032153 GDHF01030803 GDHF01027447 GDHF01017892 GDHF01017040 GDHF01017006 GDHF01012972 GDHF01007921 GDHF01005446 GDHF01004410 GDHF01002186 JAI20161.1 JAI21511.1 JAI24867.1 JAI34422.1 JAI35274.1 JAI35308.1 JAI39342.1 JAI44393.1 JAI46868.1 JAI47904.1 JAI50128.1 GAMC01005215 JAC01341.1 KQ971307 EFA11215.1 GDIQ01018587 JAN76150.1 KRT04896.1 JRES01000824 KNC28081.1 CM000157 EDW88344.1 AE014134 AY060305 AAF53325.1 AAL25344.1 CH480818 EDW51668.1 CM000361 CM002910 EDX04958.1 KMY90151.1 CH954177 EDV58646.1 AAN10845.3 CH902624 EDV33120.1 GDKW01000745 JAI55850.1 ACPB03002857 GAHY01000600 JAA76910.1 KMY90152.1 AHN54422.1 GBRD01016900 GDHC01015033 JAG48927.1 JAQ03596.1 KRJ97711.1 KQS70466.1 KPU74310.1 CH940654 EDW57529.1 CH933807 EDW13481.1 GECZ01004383 JAS65386.1 KRF77809.1 GDIP01203890 JAJ19512.1 KRG03779.1 GDRN01043299 JAI66987.1 QOIP01000003 RLU25272.1 BABH01007429 CCAG010014870 GDIQ01054124 JAN40613.1 KK852921 KDR13771.1 JXJN01027681 GL438820 EFN68414.1 GDIP01246508 JAI76893.1 AJWK01023182 AJWK01023183 AJWK01023184 KZ288430 PBC25840.1 GAPW01003855 JAC09743.1 GGFM01005558 MBW26309.1 KK107139 EZA57689.1 NNAY01001624 OXU23396.1 PYGN01001031 PSN38301.1 CH899824 EAT32454.1 APCN01005845 CH477433 EAT40999.1 EAA15121.4 AXCN02000628
KQ461187 KPJ07346.1 DS235430 EEB15691.1 LJIJ01000065 ODN03740.1 GEZM01011141 GEZM01011140 JAV93638.1 GEBQ01028915 JAT11062.1 APGK01039146 KB740967 KB632399 ENN76864.1 ERL94648.1 LJIG01022567 KRT79888.1 GFDF01001463 JAV12621.1 AAZX01003471 AAZX01016222 AAZX01018366 KQ978324 KYM95101.1 GEDC01025354 JAS11944.1 GBYB01008499 GBYB01008502 JAG78266.1 JAG78269.1 ODYU01006498 SOQ48386.1 KQ434879 KZC09966.1 UFQS01000752 UFQT01000752 SSX06634.1 SSX26981.1 GFDL01003980 JAV31065.1 GGFL01005214 MBW69392.1 GGFK01006048 MBW39369.1 CH963913 EDW77246.1 LNIX01000001 OXA62600.1 GGFL01005215 MBW69393.1 GL732573 EFX75730.1 CVRI01000058 CRL02671.1 KQ760118 OAD61917.1 CP012523 ALC40198.1 GBXI01011969 GBXI01004731 JAD02323.1 JAD09561.1 AAAB01008984 ADTU01023732 ADTU01023733 ADTU01023734 ADTU01023735 CH379061 EAL33314.1 KRT04895.1 CH479195 EDW27312.1 GBXI01014488 JAC99803.1 OUUW01000010 SPP85461.1 GAMC01005214 GAMC01005213 JAC01343.1 GAKP01006278 GAKP01006277 GAKP01006276 JAC52676.1 GDHF01032153 GDHF01030803 GDHF01027447 GDHF01017892 GDHF01017040 GDHF01017006 GDHF01012972 GDHF01007921 GDHF01005446 GDHF01004410 GDHF01002186 JAI20161.1 JAI21511.1 JAI24867.1 JAI34422.1 JAI35274.1 JAI35308.1 JAI39342.1 JAI44393.1 JAI46868.1 JAI47904.1 JAI50128.1 GAMC01005215 JAC01341.1 KQ971307 EFA11215.1 GDIQ01018587 JAN76150.1 KRT04896.1 JRES01000824 KNC28081.1 CM000157 EDW88344.1 AE014134 AY060305 AAF53325.1 AAL25344.1 CH480818 EDW51668.1 CM000361 CM002910 EDX04958.1 KMY90151.1 CH954177 EDV58646.1 AAN10845.3 CH902624 EDV33120.1 GDKW01000745 JAI55850.1 ACPB03002857 GAHY01000600 JAA76910.1 KMY90152.1 AHN54422.1 GBRD01016900 GDHC01015033 JAG48927.1 JAQ03596.1 KRJ97711.1 KQS70466.1 KPU74310.1 CH940654 EDW57529.1 CH933807 EDW13481.1 GECZ01004383 JAS65386.1 KRF77809.1 GDIP01203890 JAJ19512.1 KRG03779.1 GDRN01043299 JAI66987.1 QOIP01000003 RLU25272.1 BABH01007429 CCAG010014870 GDIQ01054124 JAN40613.1 KK852921 KDR13771.1 JXJN01027681 GL438820 EFN68414.1 GDIP01246508 JAI76893.1 AJWK01023182 AJWK01023183 AJWK01023184 KZ288430 PBC25840.1 GAPW01003855 JAC09743.1 GGFM01005558 MBW26309.1 KK107139 EZA57689.1 NNAY01001624 OXU23396.1 PYGN01001031 PSN38301.1 CH899824 EAT32454.1 APCN01005845 CH477433 EAT40999.1 EAA15121.4 AXCN02000628
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000009046
UP000094527
+ More
UP000019118 UP000030742 UP000005203 UP000002358 UP000078542 UP000076502 UP000075880 UP000007798 UP000198287 UP000075900 UP000000305 UP000183832 UP000092553 UP000005205 UP000001819 UP000008744 UP000268350 UP000007266 UP000037069 UP000091820 UP000095300 UP000095301 UP000002282 UP000000803 UP000001292 UP000000304 UP000008711 UP000007801 UP000192221 UP000015103 UP000008792 UP000009192 UP000092445 UP000279307 UP000005204 UP000092444 UP000027135 UP000092460 UP000000311 UP000092461 UP000242457 UP000053097 UP000215335 UP000245037 UP000008820 UP000075903 UP000075840 UP000075920 UP000076407 UP000075885 UP000007062 UP000075886
UP000019118 UP000030742 UP000005203 UP000002358 UP000078542 UP000076502 UP000075880 UP000007798 UP000198287 UP000075900 UP000000305 UP000183832 UP000092553 UP000005205 UP000001819 UP000008744 UP000268350 UP000007266 UP000037069 UP000091820 UP000095300 UP000095301 UP000002282 UP000000803 UP000001292 UP000000304 UP000008711 UP000007801 UP000192221 UP000015103 UP000008792 UP000009192 UP000092445 UP000279307 UP000005204 UP000092444 UP000027135 UP000092460 UP000000311 UP000092461 UP000242457 UP000053097 UP000215335 UP000245037 UP000008820 UP000075903 UP000075840 UP000075920 UP000076407 UP000075885 UP000007062 UP000075886
Interpro
SUPFAM
SSF74650
SSF74650
Gene 3D
ProteinModelPortal
A0A2A4JGU4
A0A212FGV0
A0A194PQT3
A0A0N1I6F9
E0VQN5
A0A1D2NEP7
+ More
A0A1Y1N837 A0A1B6KHY0 N6T9D7 A0A0T6AXF4 A0A087ZS11 A0A1L8E1N7 K7J8C0 A0A151I8W3 A0A1B6CES6 A0A0C9R6S1 A0A2H1W608 A0A154PDI4 A0A336MCB6 A0A182JN94 A0A1Q3FU28 A0A2M4CVM5 A0A2M4AF02 B4MYQ7 A0A226EZP0 A0A182RWF3 A0A2M4CVN3 E9GXW6 A0A1J1IR13 A0A310SGX2 A0A0M5IZA1 A0A0A1XF52 A0A1S4GZX6 A0A158NR61 Q29K64 B4GWY5 A0A0A1WLB8 A0A3B0KJY9 W8BQK5 A0A034WAT8 A0A0K8UDW8 W8BJ73 D6W7E6 A0A0P6HK98 A0A0R3P0T1 A0A0L0C762 A0A1A9WU95 A0A1I8P4I9 A0A1I8NBW9 B4P3P3 Q9V3D1 B4HX88 B4Q552 B3NAE4 Q8IP67 B3MV55 A0A1W4V7F1 A0A0P4VZ16 R4G5G2 A0A0J9R1L5 X2JAD3 A0A0K8S6M4 A0A0R1DKC4 A0A0Q5WM37 A0A1W4UV42 A0A1W4V8S6 A0A0P8Y5D5 B4M8K3 B4KIW8 A0A1B6GSJ5 A0A0Q9W7C9 A0A0P5AHW8 A0A0Q9XJF1 A0A0P4WIR6 A0A1B0A4J8 A0A3L8DYA6 H9J273 A0A1B0FFA6 A0A0N8E7J3 A0A1I8NBY4 A0A067QUV3 A0A1B0C763 E2ADQ2 A0A0P4X4F1 A0A1B0CQ74 A0A2A3E2B9 A0A023EKN2 A0A2M3ZCU3 A0A026WRG9 A0A232EY56 A0A2P8Y235 Q1DH24 A0A182VNL7 A0A2C9GRI6 Q172P9 A0A182VSJ6 A0A182X882 A0A182P2W2 Q7PW99 A0A182QZQ3
A0A1Y1N837 A0A1B6KHY0 N6T9D7 A0A0T6AXF4 A0A087ZS11 A0A1L8E1N7 K7J8C0 A0A151I8W3 A0A1B6CES6 A0A0C9R6S1 A0A2H1W608 A0A154PDI4 A0A336MCB6 A0A182JN94 A0A1Q3FU28 A0A2M4CVM5 A0A2M4AF02 B4MYQ7 A0A226EZP0 A0A182RWF3 A0A2M4CVN3 E9GXW6 A0A1J1IR13 A0A310SGX2 A0A0M5IZA1 A0A0A1XF52 A0A1S4GZX6 A0A158NR61 Q29K64 B4GWY5 A0A0A1WLB8 A0A3B0KJY9 W8BQK5 A0A034WAT8 A0A0K8UDW8 W8BJ73 D6W7E6 A0A0P6HK98 A0A0R3P0T1 A0A0L0C762 A0A1A9WU95 A0A1I8P4I9 A0A1I8NBW9 B4P3P3 Q9V3D1 B4HX88 B4Q552 B3NAE4 Q8IP67 B3MV55 A0A1W4V7F1 A0A0P4VZ16 R4G5G2 A0A0J9R1L5 X2JAD3 A0A0K8S6M4 A0A0R1DKC4 A0A0Q5WM37 A0A1W4UV42 A0A1W4V8S6 A0A0P8Y5D5 B4M8K3 B4KIW8 A0A1B6GSJ5 A0A0Q9W7C9 A0A0P5AHW8 A0A0Q9XJF1 A0A0P4WIR6 A0A1B0A4J8 A0A3L8DYA6 H9J273 A0A1B0FFA6 A0A0N8E7J3 A0A1I8NBY4 A0A067QUV3 A0A1B0C763 E2ADQ2 A0A0P4X4F1 A0A1B0CQ74 A0A2A3E2B9 A0A023EKN2 A0A2M3ZCU3 A0A026WRG9 A0A232EY56 A0A2P8Y235 Q1DH24 A0A182VNL7 A0A2C9GRI6 Q172P9 A0A182VSJ6 A0A182X882 A0A182P2W2 Q7PW99 A0A182QZQ3
PDB
2CIS
E-value=1.58971e-36,
Score=381
Ontologies
PATHWAY
GO
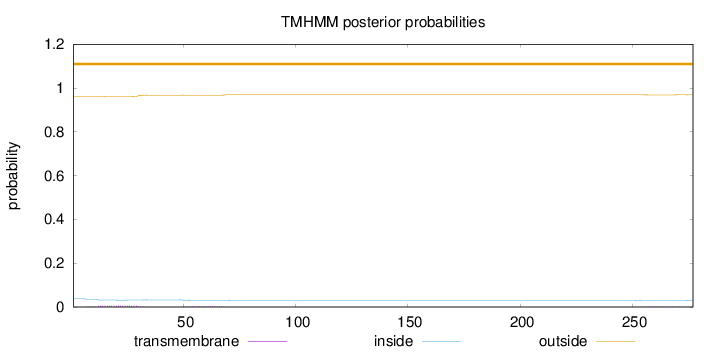
Topology
Length:
277
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22428
Exp number, first 60 AAs:
0.16855
Total prob of N-in:
0.03728
outside
1 - 277
Population Genetic Test Statistics
Pi
241.844575
Theta
177.930587
Tajima's D
1.262326
CLR
0.181254
CSRT
0.729663516824159
Interpretation
Uncertain
