Gene
KWMTBOMO03000 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003586
Annotation
PREDICTED:_28S_ribosomal_protein_S31?_mitochondrial_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.58
Sequence
CDS
ATGTTAACAAGACTCAAGTTCAGAAAAGATTTAAGGGTAATCGCACGGTGTTTGTCAGACAAATCCAAGGATAATGATGGGAAGCCTCAAGTAATTCAAGATGCCAAGAAGAAAACCGTAAAAAATGATCCGGCTACTGAAAAGATTCAAGAACTGTTGAAAAGCATGATGGCACCTCCAAAAATAAGCGAAGCTGAGTACAGAGAAAAATTCGCAACATCTCCAGATACTAGGCGAAGTAAACAAGTGGACGAAATAGAAGTGAAGACTGAAAAAATAGAGGAGAGCATCACAAAAGCTGCTACTGATGTTGCTCAAGCTATAGGGGGTGATGTCAAACAAACAGAAGCAGAGCTTCTATCTCGTGTACTTGGGAAAATCAATCAAACTTCTACCACTCTCAGTGACCTTTTAGTTGGGATGAAGGTAGACAGAACAGTAGAACCTGATGAAGTCCAAAAAAAACCAGATACTAGAGAGCAGCAAGTGAAAAGACTTGTAAGCAAAGCAAAGACAACTGAAGCTTCTCCTACTAGATATTCACAACGGAAATCAGCATATGTTCCCAATGACAGGACAAGGCAAGGAAGAGACAGTAACAGAAGTGGTACTCCACAAATCACAGAAATTGATATATTTGGTGGTGAGCCACTTGGTATATTCAAGACAACAGAACCAAACTATGGAACAAAACTGGATGTTTGGGAGAACCTAAAAAAACGAGAGCTAAGTTTAGCAACTGCACAGCCCCCGGCCAATTATTTCCAGAAAATTATCCTGTGGACCAATCAGGGTAAAGTTTGGAAATTCCCCATTGACAACGAGCAAGGGCTGGAAGAGGAACAAAAGATACATTTCTCCGAGCACATATTTTTGGACACACATCTAGAAGACTGGTGCCCGAAGAAAGGACCGATACGCCACTTCATGGAGCTCGTCTGCGTCGGACTCTCAAAGAATCCATTTTACACCGCCCAGGAGAAAAAAGAACACATCATGTGGTACAAAGACTATTTTGTAACGAAAAAGGATTTATTGACCGAAGTAGGAGCTTGGGATCTACCTAACAGCAACAAAGAGGCGTCGGTCTAA
Protein
MLTRLKFRKDLRVIARCLSDKSKDNDGKPQVIQDAKKKTVKNDPATEKIQELLKSMMAPPKISEAEYREKFATSPDTRRSKQVDEIEVKTEKIEESITKAATDVAQAIGGDVKQTEAELLSRVLGKINQTSTTLSDLLVGMKVDRTVEPDEVQKKPDTREQQVKRLVSKAKTTEASPTRYSQRKSAYVPNDRTRQGRDSNRSGTPQITEIDIFGGEPLGIFKTTEPNYGTKLDVWENLKKRELSLATAQPPANYFQKIILWTNQGKVWKFPIDNEQGLEEEQKIHFSEHIFLDTHLEDWCPKKGPIRHFMELVCVGLSKNPFYTAQEKKEHIMWYKDYFVTKKDLLTEVGAWDLPNSNKEASV
Summary
Description
Seems to be required for maximal rate of protein biosynthesis. Enhances ribosome dissociation into subunits and stabilizes the binding of the initiator Met-tRNA(I) to 40 S ribosomal subunits.
Similarity
Belongs to the tumor necrosis factor family.
Belongs to the eIF-1A family.
Belongs to the eIF-1A family.
Uniprot
H9J249
A0A2H1W5T2
A0A2A4JFP0
A0A212FGX7
A0A0N1ID54
A0A194PQ63
+ More
A0A3B0JSX2 A0A1W4WL93 Q9VUX1 Q95S88 B4HIM5 D6WRW9 B4QLS6 Q29D95 A0A1W4VMQ6 B4ITV3 B4H7T1 B3M9T7 A0A1S4FUL9 Q16NN5 A0A2J7PF71 V5GTE1 B3NDQ3 A0A067RLY8 A0A1Q3F7T8 A0A182FB78 W5JWZ5 A0A182Y9I1 A0A182QVY6 A0A1Y1LIN9 A0A182RVG5 T1DRM6 A0A182ISX8 B4IYF8 A0A182H366 A0A0K8W8G9 B0WW32 A0A0V0G7E2 Q7PZI5 A0A182SMT9 B4LG31 A0A034VF70 A0A182WE11 J9JND0 B4N729 A0A182L565 A0A2S2NMK0 A0A1B0CK83 A0A1S4H7V3 A0A182HHM0 A0A2S2QWC8 A0A2H8TSA4 A0A084WII3 A0A182PCA0 W8BQ63 A0A182XJR3 A0A0L7L103 A0A182N2S8 A0A182VBH5 A0A182M6N4 A0A182TNC0 A0A1I8NU28 U5ESY9 A0A0A9Z6Q4 A0A182JPY0 A0A0A1XRU9 A0A026WNK4 T1PG90 B4KYG4 A0A1B6IPQ4 A0A1J1HKJ6 A0A1B6GL32 A0A146M7M0 A0A1D2NMA1 A0A1L8E0A9 A0A151J9H9 A0A1L8E042 A0A0P4VMF6 A0A232F936 K7J1N7 A0A1B6LRN0 A0A1A9V584 A0A158NZ52 D3TMC7 A0A1B0AD54 A0A151WWS9 A0A195CW05 E2A636 A0A1A9YIL7 A0A0L0CJT6 A0A1B0D3Q2 A0A1A9WDH7 A0A1B0AQR0 A0A226DP35 A0A195F7C2 A0A2R7WMY3 A0A293LF05
A0A3B0JSX2 A0A1W4WL93 Q9VUX1 Q95S88 B4HIM5 D6WRW9 B4QLS6 Q29D95 A0A1W4VMQ6 B4ITV3 B4H7T1 B3M9T7 A0A1S4FUL9 Q16NN5 A0A2J7PF71 V5GTE1 B3NDQ3 A0A067RLY8 A0A1Q3F7T8 A0A182FB78 W5JWZ5 A0A182Y9I1 A0A182QVY6 A0A1Y1LIN9 A0A182RVG5 T1DRM6 A0A182ISX8 B4IYF8 A0A182H366 A0A0K8W8G9 B0WW32 A0A0V0G7E2 Q7PZI5 A0A182SMT9 B4LG31 A0A034VF70 A0A182WE11 J9JND0 B4N729 A0A182L565 A0A2S2NMK0 A0A1B0CK83 A0A1S4H7V3 A0A182HHM0 A0A2S2QWC8 A0A2H8TSA4 A0A084WII3 A0A182PCA0 W8BQ63 A0A182XJR3 A0A0L7L103 A0A182N2S8 A0A182VBH5 A0A182M6N4 A0A182TNC0 A0A1I8NU28 U5ESY9 A0A0A9Z6Q4 A0A182JPY0 A0A0A1XRU9 A0A026WNK4 T1PG90 B4KYG4 A0A1B6IPQ4 A0A1J1HKJ6 A0A1B6GL32 A0A146M7M0 A0A1D2NMA1 A0A1L8E0A9 A0A151J9H9 A0A1L8E042 A0A0P4VMF6 A0A232F936 K7J1N7 A0A1B6LRN0 A0A1A9V584 A0A158NZ52 D3TMC7 A0A1B0AD54 A0A151WWS9 A0A195CW05 E2A636 A0A1A9YIL7 A0A0L0CJT6 A0A1B0D3Q2 A0A1A9WDH7 A0A1B0AQR0 A0A226DP35 A0A195F7C2 A0A2R7WMY3 A0A293LF05
Pubmed
19121390
22118469
26354079
10731132
12537568
12537572
+ More
12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 18362917 19820115 22936249 15632085 17550304 17510324 24845553 20920257 23761445 25244985 28004739 26483478 12364791 25348373 20966253 24438588 24495485 26227816 25401762 25830018 24508170 30249741 25315136 26823975 27289101 27129103 28648823 20075255 21347285 20353571 20798317 26108605
12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 18362917 19820115 22936249 15632085 17550304 17510324 24845553 20920257 23761445 25244985 28004739 26483478 12364791 25348373 20966253 24438588 24495485 26227816 25401762 25830018 24508170 30249741 25315136 26823975 27289101 27129103 28648823 20075255 21347285 20353571 20798317 26108605
EMBL
BABH01007429
ODYU01006498
SOQ48387.1
NWSH01001631
PCG70639.1
AGBW02008594
+ More
OWR52963.1 KQ461187 KPJ07345.1 KQ459597 KPI95103.1 OUUW01000002 SPP76799.1 AE014296 AY089653 AAF49552.1 AAL90391.1 AY060911 AAL28459.1 CH480815 EDW41655.1 KQ971351 EFA06593.1 CM000363 CM002912 EDX10625.1 KMY99894.1 CH379070 EAL30519.1 CH891734 EDW99816.1 CH479219 EDW34721.1 CH902618 EDV40128.1 CH477816 EAT35950.1 NEVH01026085 PNF14974.1 GALX01001047 JAB67419.1 CH954178 EDV52186.2 KK852595 KDR20607.1 GFDL01011472 JAV23573.1 ADMH02000120 ETN67714.1 AXCN02001087 GEZM01054675 JAV73493.1 GAMD01001864 JAA99726.1 CH916366 EDV97631.1 JXUM01106761 KQ565082 KXJ71360.1 GDHF01004816 JAI47498.1 DS232137 EDS35864.1 GECL01002222 JAP03902.1 AAAB01008986 EAA00689.4 CH940647 EDW70430.1 GAKP01018769 JAC40183.1 ABLF02018654 CH964168 EDW80168.1 GGMR01005739 MBY18358.1 AJWK01015894 APCN01002428 GGMS01012866 MBY82069.1 GFXV01005262 GFXV01006007 MBW17067.1 MBW17812.1 ATLV01023934 KE525347 KFB50027.1 GAMC01005388 JAC01168.1 JTDY01003715 KOB69105.1 AXCM01001233 GANO01003003 JAB56868.1 GBHO01004561 GBRD01008465 JAG39043.1 JAG57356.1 GBXI01000662 JAD13630.1 KK107151 QOIP01000003 EZA57251.1 RLU24390.1 KA647827 AFP62456.1 CH933809 EDW18775.1 GECU01018826 JAS88880.1 CVRI01000006 CRK88050.1 GECZ01006613 JAS63156.1 GDHC01003200 JAQ15429.1 LJIJ01000006 ODN06408.1 GFDF01001988 JAV12096.1 KQ979403 KYN21725.1 GFDF01001971 JAV12113.1 GDKW01002624 JAI53971.1 NNAY01000619 OXU27364.1 GEBQ01013589 JAT26388.1 ADTU01003818 CCAG010002173 EZ422579 ADD18855.1 KQ982684 KYQ52343.1 KQ977220 KYN04815.1 GL437108 EFN71095.1 JRES01000303 KNC32510.1 AJVK01011006 JXJN01002008 LNIX01000015 OXA46594.1 KQ981768 KYN35969.1 KK854934 PTY19785.1 GFWV01002592 MAA27322.1
OWR52963.1 KQ461187 KPJ07345.1 KQ459597 KPI95103.1 OUUW01000002 SPP76799.1 AE014296 AY089653 AAF49552.1 AAL90391.1 AY060911 AAL28459.1 CH480815 EDW41655.1 KQ971351 EFA06593.1 CM000363 CM002912 EDX10625.1 KMY99894.1 CH379070 EAL30519.1 CH891734 EDW99816.1 CH479219 EDW34721.1 CH902618 EDV40128.1 CH477816 EAT35950.1 NEVH01026085 PNF14974.1 GALX01001047 JAB67419.1 CH954178 EDV52186.2 KK852595 KDR20607.1 GFDL01011472 JAV23573.1 ADMH02000120 ETN67714.1 AXCN02001087 GEZM01054675 JAV73493.1 GAMD01001864 JAA99726.1 CH916366 EDV97631.1 JXUM01106761 KQ565082 KXJ71360.1 GDHF01004816 JAI47498.1 DS232137 EDS35864.1 GECL01002222 JAP03902.1 AAAB01008986 EAA00689.4 CH940647 EDW70430.1 GAKP01018769 JAC40183.1 ABLF02018654 CH964168 EDW80168.1 GGMR01005739 MBY18358.1 AJWK01015894 APCN01002428 GGMS01012866 MBY82069.1 GFXV01005262 GFXV01006007 MBW17067.1 MBW17812.1 ATLV01023934 KE525347 KFB50027.1 GAMC01005388 JAC01168.1 JTDY01003715 KOB69105.1 AXCM01001233 GANO01003003 JAB56868.1 GBHO01004561 GBRD01008465 JAG39043.1 JAG57356.1 GBXI01000662 JAD13630.1 KK107151 QOIP01000003 EZA57251.1 RLU24390.1 KA647827 AFP62456.1 CH933809 EDW18775.1 GECU01018826 JAS88880.1 CVRI01000006 CRK88050.1 GECZ01006613 JAS63156.1 GDHC01003200 JAQ15429.1 LJIJ01000006 ODN06408.1 GFDF01001988 JAV12096.1 KQ979403 KYN21725.1 GFDF01001971 JAV12113.1 GDKW01002624 JAI53971.1 NNAY01000619 OXU27364.1 GEBQ01013589 JAT26388.1 ADTU01003818 CCAG010002173 EZ422579 ADD18855.1 KQ982684 KYQ52343.1 KQ977220 KYN04815.1 GL437108 EFN71095.1 JRES01000303 KNC32510.1 AJVK01011006 JXJN01002008 LNIX01000015 OXA46594.1 KQ981768 KYN35969.1 KK854934 PTY19785.1 GFWV01002592 MAA27322.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000268350
+ More
UP000192223 UP000000803 UP000001292 UP000007266 UP000000304 UP000001819 UP000192221 UP000002282 UP000008744 UP000007801 UP000008820 UP000235965 UP000008711 UP000027135 UP000069272 UP000000673 UP000076408 UP000075886 UP000075900 UP000075880 UP000001070 UP000069940 UP000249989 UP000002320 UP000007062 UP000075901 UP000008792 UP000075920 UP000007819 UP000007798 UP000075882 UP000092461 UP000075840 UP000030765 UP000075885 UP000076407 UP000037510 UP000075884 UP000075903 UP000075883 UP000075902 UP000095300 UP000075881 UP000053097 UP000279307 UP000095301 UP000009192 UP000183832 UP000094527 UP000078492 UP000215335 UP000002358 UP000078200 UP000005205 UP000092444 UP000092445 UP000075809 UP000078542 UP000000311 UP000092443 UP000037069 UP000092462 UP000091820 UP000092460 UP000198287 UP000078541
UP000192223 UP000000803 UP000001292 UP000007266 UP000000304 UP000001819 UP000192221 UP000002282 UP000008744 UP000007801 UP000008820 UP000235965 UP000008711 UP000027135 UP000069272 UP000000673 UP000076408 UP000075886 UP000075900 UP000075880 UP000001070 UP000069940 UP000249989 UP000002320 UP000007062 UP000075901 UP000008792 UP000075920 UP000007819 UP000007798 UP000075882 UP000092461 UP000075840 UP000030765 UP000075885 UP000076407 UP000037510 UP000075884 UP000075903 UP000075883 UP000075902 UP000095300 UP000075881 UP000053097 UP000279307 UP000095301 UP000009192 UP000183832 UP000094527 UP000078492 UP000215335 UP000002358 UP000078200 UP000005205 UP000092444 UP000092445 UP000075809 UP000078542 UP000000311 UP000092443 UP000037069 UP000092462 UP000091820 UP000092460 UP000198287 UP000078541
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J249
A0A2H1W5T2
A0A2A4JFP0
A0A212FGX7
A0A0N1ID54
A0A194PQ63
+ More
A0A3B0JSX2 A0A1W4WL93 Q9VUX1 Q95S88 B4HIM5 D6WRW9 B4QLS6 Q29D95 A0A1W4VMQ6 B4ITV3 B4H7T1 B3M9T7 A0A1S4FUL9 Q16NN5 A0A2J7PF71 V5GTE1 B3NDQ3 A0A067RLY8 A0A1Q3F7T8 A0A182FB78 W5JWZ5 A0A182Y9I1 A0A182QVY6 A0A1Y1LIN9 A0A182RVG5 T1DRM6 A0A182ISX8 B4IYF8 A0A182H366 A0A0K8W8G9 B0WW32 A0A0V0G7E2 Q7PZI5 A0A182SMT9 B4LG31 A0A034VF70 A0A182WE11 J9JND0 B4N729 A0A182L565 A0A2S2NMK0 A0A1B0CK83 A0A1S4H7V3 A0A182HHM0 A0A2S2QWC8 A0A2H8TSA4 A0A084WII3 A0A182PCA0 W8BQ63 A0A182XJR3 A0A0L7L103 A0A182N2S8 A0A182VBH5 A0A182M6N4 A0A182TNC0 A0A1I8NU28 U5ESY9 A0A0A9Z6Q4 A0A182JPY0 A0A0A1XRU9 A0A026WNK4 T1PG90 B4KYG4 A0A1B6IPQ4 A0A1J1HKJ6 A0A1B6GL32 A0A146M7M0 A0A1D2NMA1 A0A1L8E0A9 A0A151J9H9 A0A1L8E042 A0A0P4VMF6 A0A232F936 K7J1N7 A0A1B6LRN0 A0A1A9V584 A0A158NZ52 D3TMC7 A0A1B0AD54 A0A151WWS9 A0A195CW05 E2A636 A0A1A9YIL7 A0A0L0CJT6 A0A1B0D3Q2 A0A1A9WDH7 A0A1B0AQR0 A0A226DP35 A0A195F7C2 A0A2R7WMY3 A0A293LF05
A0A3B0JSX2 A0A1W4WL93 Q9VUX1 Q95S88 B4HIM5 D6WRW9 B4QLS6 Q29D95 A0A1W4VMQ6 B4ITV3 B4H7T1 B3M9T7 A0A1S4FUL9 Q16NN5 A0A2J7PF71 V5GTE1 B3NDQ3 A0A067RLY8 A0A1Q3F7T8 A0A182FB78 W5JWZ5 A0A182Y9I1 A0A182QVY6 A0A1Y1LIN9 A0A182RVG5 T1DRM6 A0A182ISX8 B4IYF8 A0A182H366 A0A0K8W8G9 B0WW32 A0A0V0G7E2 Q7PZI5 A0A182SMT9 B4LG31 A0A034VF70 A0A182WE11 J9JND0 B4N729 A0A182L565 A0A2S2NMK0 A0A1B0CK83 A0A1S4H7V3 A0A182HHM0 A0A2S2QWC8 A0A2H8TSA4 A0A084WII3 A0A182PCA0 W8BQ63 A0A182XJR3 A0A0L7L103 A0A182N2S8 A0A182VBH5 A0A182M6N4 A0A182TNC0 A0A1I8NU28 U5ESY9 A0A0A9Z6Q4 A0A182JPY0 A0A0A1XRU9 A0A026WNK4 T1PG90 B4KYG4 A0A1B6IPQ4 A0A1J1HKJ6 A0A1B6GL32 A0A146M7M0 A0A1D2NMA1 A0A1L8E0A9 A0A151J9H9 A0A1L8E042 A0A0P4VMF6 A0A232F936 K7J1N7 A0A1B6LRN0 A0A1A9V584 A0A158NZ52 D3TMC7 A0A1B0AD54 A0A151WWS9 A0A195CW05 E2A636 A0A1A9YIL7 A0A0L0CJT6 A0A1B0D3Q2 A0A1A9WDH7 A0A1B0AQR0 A0A226DP35 A0A195F7C2 A0A2R7WMY3 A0A293LF05
PDB
6NF8
E-value=8.18061e-30,
Score=325
Ontologies
GO
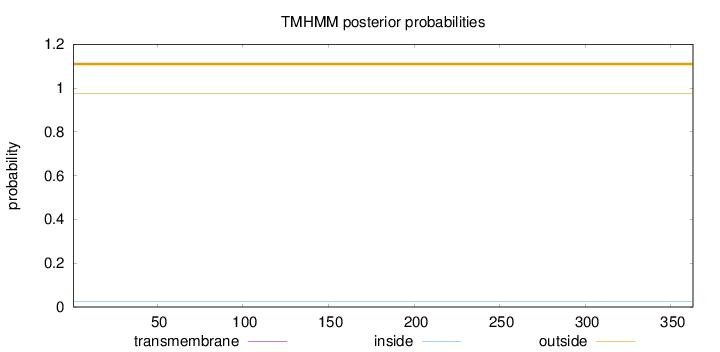
Topology
Length:
363
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00057
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02584
outside
1 - 363
Population Genetic Test Statistics
Pi
338.57327
Theta
183.72779
Tajima's D
2.473907
CLR
0.488494
CSRT
0.942602869856507
Interpretation
Uncertain
