Gene
KWMTBOMO02996
Pre Gene Modal
BGIBMGA003616
Annotation
PREDICTED:_phospholipid_scramblase_2_[Bombyx_mori]
Full name
Phospholipid scramblase
Location in the cell
Extracellular Reliability : 1.183 PlasmaMembrane Reliability : 1.233
Sequence
CDS
ATGGATCTGAAAGTATTTGATCGTGAAACTGGAGTGGGCGATCGAGGAATCGATAATCCAGCTGCCACAGACGATTATGGATCCCACGTAAGACCTGATTTGGTAATTAGCGTTCAGCCGCAATCACATGGAACTCGTGCAGAATTACCAGTGACTACGGTCAACTGGCGCTCCAACCCTCGAGGTCAGTTCATCCCGGTACATGGATTGGACTTTCTAATTGGAGTTTCCAGTCTTCTAATACAACAGACCGTTGAGCTTGATGATCTGACTTCAAAAATAGAATCGGAGAATCGTTACATAGTGAGGGTACCGAATGGAGAAGCTTTGTACATCGGCAACGAAGTGTCGTGTCCAACTCAGAGATGTATCTGTGGGAATGGTCGAGCCTTTATGATGCATCTTCATGACAACACGAGGCAAGAGGCCATGGTGCTGAAGCGAAGGTTGGCAGGGGCATCTTGTTTGTGGCCTTGTCGACTGCAAGAAATGAAAGTGATAACGCCTCCAGGCGACTACGTGGGACGTATCCAACAAAGCTGTACATGGATGATACCAGAGTACAGGGTTAGGGACGCCAACGACCAGACCCTGTTCATTATTGAAGGTCCAGCCGTATTGAAGAGAAGTGCTTTGATGTTATCCGAGTTTAAGATTCTGACTGCAGATTCCTCGAGAATCGTGGGACGGATAGCCCATGGCTGGGATCGTAATCTCGTCAGCTTCGCGACAACTCTCCAAATCCCTGATACAGCTGTACAACCTAAACATAAAGCATTGCTACTGGCCGCTACGTTTTTATTGGAGTACACATATTTTGAAAGAACAAAATCGAGTTGTACGAGATGCCTTTTTTGCTGA
Protein
MDLKVFDRETGVGDRGIDNPAATDDYGSHVRPDLVISVQPQSHGTRAELPVTTVNWRSNPRGQFIPVHGLDFLIGVSSLLIQQTVELDDLTSKIESENRYIVRVPNGEALYIGNEVSCPTQRCICGNGRAFMMHLHDNTRQEAMVLKRRLAGASCLWPCRLQEMKVITPPGDYVGRIQQSCTWMIPEYRVRDANDQTLFIIEGPAVLKRSALMLSEFKILTADSSRIVGRIAHGWDRNLVSFATTLQIPDTAVQPKHKALLLAATFLLEYTYFERTKSSCTRCLFC
Summary
Description
May mediate accelerated ATP-independent bidirectional transbilayer migration of phospholipids upon binding calcium ions that results in a loss of phospholipid asymmetry in the plasma membrane.
Cofactor
Ca(2+)
Similarity
Belongs to the phospholipid scramblase family.
Uniprot
A0A2A4JGQ3
A0A1E1VZT3
A0A2H1W9H4
A0A194PP65
A0A212EVF7
H9J279
+ More
A0A0N0PAM0 A0A1B6J2B1 A0A1B6DVY3 A0A2J7QX24 A0A2J7QX19 V5GLD7 V5GX66 A0A1Y1NF14 A0A139WNT6 D6W758 A0A023FBA2 A0A067RC55 A0A224XHI0 A0A146LE77 A0A146KLW3 A0A2P8Y3W2 J3JUV8 N6T2A2 B3MEQ8 B4P7K7 A0A023FB97 B4J846 B3NSN2 B4MK84 A0A1W4U5M6 B4KQ27 Q28WS6 B4H6B7 A1Z8F5 B4ME12 A0A3B0JL62 Q8SZ85 B5RJ61 A0A0A1XPE8 A0A0Q9WBG8 A0A0L0BP84 A0A3B0J034 B4HN40 W8BRW9 R4WR60 A0A1L8EGK8 W8BED8 A0A0K8W9F6 A0A1I8PH54 A0A0Q9WP14 A0A034VDF6 A0A0K8UKJ5 A0A1I8NAH3 B4QB34 A0A0B4KEL2 A0A034V927 W8C1R3 A0A0N7ZXB1 A0A336LV26 E9GIA0 A0A0P5HUT3 W5J5J0 A0A0P5T9E0 A0A0P5A6Q2 A0A1Y1JWZ4 A0A067QG92 C3ZIU3 A0A182YJI1 A0A182SMD3 A0A1W4WF47 A0A1W4WQ33 A0A1B6HDV9 A0A1B6LRE2 A0A0P6CRD8 A0A0P6CLZ2 A0A182L446 A0A0P5SL04 A0A182R9A9 A0A060YVX0 A0A1L8E3L1 A0A1B6LTY2 T1IB30 A0A182X243 A0A182N7I8 A0A182I6A4 E0VXM0 A0A1S3KKV9 A0A0A9XAE0 A0A182VGT9 A0A2J7R040 A0A182QYG9 A0A2J7R027 A0A2D0R9Y6 A0A2J7R038 A0A1B6E985
A0A0N0PAM0 A0A1B6J2B1 A0A1B6DVY3 A0A2J7QX24 A0A2J7QX19 V5GLD7 V5GX66 A0A1Y1NF14 A0A139WNT6 D6W758 A0A023FBA2 A0A067RC55 A0A224XHI0 A0A146LE77 A0A146KLW3 A0A2P8Y3W2 J3JUV8 N6T2A2 B3MEQ8 B4P7K7 A0A023FB97 B4J846 B3NSN2 B4MK84 A0A1W4U5M6 B4KQ27 Q28WS6 B4H6B7 A1Z8F5 B4ME12 A0A3B0JL62 Q8SZ85 B5RJ61 A0A0A1XPE8 A0A0Q9WBG8 A0A0L0BP84 A0A3B0J034 B4HN40 W8BRW9 R4WR60 A0A1L8EGK8 W8BED8 A0A0K8W9F6 A0A1I8PH54 A0A0Q9WP14 A0A034VDF6 A0A0K8UKJ5 A0A1I8NAH3 B4QB34 A0A0B4KEL2 A0A034V927 W8C1R3 A0A0N7ZXB1 A0A336LV26 E9GIA0 A0A0P5HUT3 W5J5J0 A0A0P5T9E0 A0A0P5A6Q2 A0A1Y1JWZ4 A0A067QG92 C3ZIU3 A0A182YJI1 A0A182SMD3 A0A1W4WF47 A0A1W4WQ33 A0A1B6HDV9 A0A1B6LRE2 A0A0P6CRD8 A0A0P6CLZ2 A0A182L446 A0A0P5SL04 A0A182R9A9 A0A060YVX0 A0A1L8E3L1 A0A1B6LTY2 T1IB30 A0A182X243 A0A182N7I8 A0A182I6A4 E0VXM0 A0A1S3KKV9 A0A0A9XAE0 A0A182VGT9 A0A2J7R040 A0A182QYG9 A0A2J7R027 A0A2D0R9Y6 A0A2J7R038 A0A1B6E985
Pubmed
26354079
22118469
19121390
28004739
18362917
19820115
+ More
25474469 24845553 26823975 29403074 22516182 23537049 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25830018 26108605 24495485 23691247 25348373 25315136 22936249 21292972 20920257 23761445 18563158 25244985 20966253 24755649 20566863 25401762
25474469 24845553 26823975 29403074 22516182 23537049 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25830018 26108605 24495485 23691247 25348373 25315136 22936249 21292972 20920257 23761445 18563158 25244985 20966253 24755649 20566863 25401762
EMBL
NWSH01001547
PCG70898.1
GDQN01010821
JAT80233.1
ODYU01007111
SOQ49596.1
+ More
KQ459597 KPI95097.1 AGBW02012195 OWR45472.1 BABH01007434 KQ461187 KPJ07340.1 GECU01014404 JAS93302.1 GEDC01020451 GEDC01007463 JAS16847.1 JAS29835.1 NEVH01009398 PNF33133.1 PNF33132.1 GALX01003562 JAB64904.1 GALX01003563 JAB64903.1 GEZM01003858 JAV96523.1 KQ971307 KYB29660.1 EFA11407.1 GBBI01000423 JAC18289.1 KK852554 KDR21466.1 GFTR01004481 JAW11945.1 GDHC01012834 JAQ05795.1 GDHC01021076 JAP97552.1 PYGN01000957 PSN38929.1 BT127023 AEE61985.1 APGK01047232 APGK01047233 KB741077 KB631792 ENN74254.1 ERL86119.1 CH902619 EDV35522.2 CM000158 EDW90042.1 GBBI01000428 JAC18284.1 CH916367 EDW01183.1 CH954179 EDV56534.1 CH963846 EDW72523.1 CH933808 EDW08129.1 CM000071 EAL26591.1 CH479213 EDW33341.1 AE013599 AAM68719.1 CH940662 EDW58777.1 OUUW01000001 SPP73976.1 AY071050 AAL48672.1 BT044335 ACH92400.1 GBXI01001849 JAD12443.1 KRF78479.1 JRES01001578 KNC21865.1 SPP73975.1 CH480816 EDW47344.1 GAMC01006907 JAB99648.1 AK418186 BAN21401.1 GFDG01001015 JAV17784.1 GAMC01006905 JAB99650.1 GDHF01004491 JAI47823.1 KRF97591.1 GAKP01019152 JAC39800.1 GDHF01025105 GDHF01023532 GDHF01018295 JAI27209.1 JAI28782.1 JAI34019.1 CM000362 CM002911 EDX06567.1 KMY92913.1 AGB93417.1 GAKP01019151 JAC39801.1 GAMC01006904 JAB99651.1 GDIP01203449 JAJ19953.1 UFQT01000126 SSX20523.1 GL732546 EFX80807.1 GDIQ01245465 LRGB01003375 JAK06260.1 KZS02915.1 ADMH02002096 ETN59256.1 GDIQ01095111 JAL56615.1 GDIP01203448 JAJ19954.1 GEZM01103143 GEZM01103142 JAV51526.1 KK853500 KDR07158.1 GG666629 EEN47544.1 GECU01034927 GECU01013232 JAS72779.1 JAS94474.1 GEBQ01013755 JAT26222.1 GDIQ01089294 JAN05443.1 GDIQ01089295 JAN05442.1 GDIQ01091689 JAL60037.1 FR915643 CDQ93644.1 GFDF01000838 JAV13246.1 GEBQ01012851 JAT27126.1 ACPB03021686 APCN01003602 DS235833 EEB18126.1 GBHO01025937 GBHO01025936 GDHC01020916 GDHC01012357 GDHC01004641 JAG17667.1 JAG17668.1 JAP97712.1 JAQ06272.1 JAQ13988.1 NEVH01008299 PNF34190.1 AXCN02000574 PNF34192.1 PNF34198.1 GEDC01002799 JAS34499.1
KQ459597 KPI95097.1 AGBW02012195 OWR45472.1 BABH01007434 KQ461187 KPJ07340.1 GECU01014404 JAS93302.1 GEDC01020451 GEDC01007463 JAS16847.1 JAS29835.1 NEVH01009398 PNF33133.1 PNF33132.1 GALX01003562 JAB64904.1 GALX01003563 JAB64903.1 GEZM01003858 JAV96523.1 KQ971307 KYB29660.1 EFA11407.1 GBBI01000423 JAC18289.1 KK852554 KDR21466.1 GFTR01004481 JAW11945.1 GDHC01012834 JAQ05795.1 GDHC01021076 JAP97552.1 PYGN01000957 PSN38929.1 BT127023 AEE61985.1 APGK01047232 APGK01047233 KB741077 KB631792 ENN74254.1 ERL86119.1 CH902619 EDV35522.2 CM000158 EDW90042.1 GBBI01000428 JAC18284.1 CH916367 EDW01183.1 CH954179 EDV56534.1 CH963846 EDW72523.1 CH933808 EDW08129.1 CM000071 EAL26591.1 CH479213 EDW33341.1 AE013599 AAM68719.1 CH940662 EDW58777.1 OUUW01000001 SPP73976.1 AY071050 AAL48672.1 BT044335 ACH92400.1 GBXI01001849 JAD12443.1 KRF78479.1 JRES01001578 KNC21865.1 SPP73975.1 CH480816 EDW47344.1 GAMC01006907 JAB99648.1 AK418186 BAN21401.1 GFDG01001015 JAV17784.1 GAMC01006905 JAB99650.1 GDHF01004491 JAI47823.1 KRF97591.1 GAKP01019152 JAC39800.1 GDHF01025105 GDHF01023532 GDHF01018295 JAI27209.1 JAI28782.1 JAI34019.1 CM000362 CM002911 EDX06567.1 KMY92913.1 AGB93417.1 GAKP01019151 JAC39801.1 GAMC01006904 JAB99651.1 GDIP01203449 JAJ19953.1 UFQT01000126 SSX20523.1 GL732546 EFX80807.1 GDIQ01245465 LRGB01003375 JAK06260.1 KZS02915.1 ADMH02002096 ETN59256.1 GDIQ01095111 JAL56615.1 GDIP01203448 JAJ19954.1 GEZM01103143 GEZM01103142 JAV51526.1 KK853500 KDR07158.1 GG666629 EEN47544.1 GECU01034927 GECU01013232 JAS72779.1 JAS94474.1 GEBQ01013755 JAT26222.1 GDIQ01089294 JAN05443.1 GDIQ01089295 JAN05442.1 GDIQ01091689 JAL60037.1 FR915643 CDQ93644.1 GFDF01000838 JAV13246.1 GEBQ01012851 JAT27126.1 ACPB03021686 APCN01003602 DS235833 EEB18126.1 GBHO01025937 GBHO01025936 GDHC01020916 GDHC01012357 GDHC01004641 JAG17667.1 JAG17668.1 JAP97712.1 JAQ06272.1 JAQ13988.1 NEVH01008299 PNF34190.1 AXCN02000574 PNF34192.1 PNF34198.1 GEDC01002799 JAS34499.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000005204
UP000053240
UP000235965
+ More
UP000007266 UP000027135 UP000245037 UP000019118 UP000030742 UP000007801 UP000002282 UP000001070 UP000008711 UP000007798 UP000192221 UP000009192 UP000001819 UP000008744 UP000000803 UP000008792 UP000268350 UP000037069 UP000001292 UP000095300 UP000095301 UP000000304 UP000000305 UP000076858 UP000000673 UP000001554 UP000076408 UP000075901 UP000192223 UP000075882 UP000075900 UP000193380 UP000015103 UP000076407 UP000075884 UP000075840 UP000009046 UP000087266 UP000075903 UP000075886 UP000221080
UP000007266 UP000027135 UP000245037 UP000019118 UP000030742 UP000007801 UP000002282 UP000001070 UP000008711 UP000007798 UP000192221 UP000009192 UP000001819 UP000008744 UP000000803 UP000008792 UP000268350 UP000037069 UP000001292 UP000095300 UP000095301 UP000000304 UP000000305 UP000076858 UP000000673 UP000001554 UP000076408 UP000075901 UP000192223 UP000075882 UP000075900 UP000193380 UP000015103 UP000076407 UP000075884 UP000075840 UP000009046 UP000087266 UP000075903 UP000075886 UP000221080
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JGQ3
A0A1E1VZT3
A0A2H1W9H4
A0A194PP65
A0A212EVF7
H9J279
+ More
A0A0N0PAM0 A0A1B6J2B1 A0A1B6DVY3 A0A2J7QX24 A0A2J7QX19 V5GLD7 V5GX66 A0A1Y1NF14 A0A139WNT6 D6W758 A0A023FBA2 A0A067RC55 A0A224XHI0 A0A146LE77 A0A146KLW3 A0A2P8Y3W2 J3JUV8 N6T2A2 B3MEQ8 B4P7K7 A0A023FB97 B4J846 B3NSN2 B4MK84 A0A1W4U5M6 B4KQ27 Q28WS6 B4H6B7 A1Z8F5 B4ME12 A0A3B0JL62 Q8SZ85 B5RJ61 A0A0A1XPE8 A0A0Q9WBG8 A0A0L0BP84 A0A3B0J034 B4HN40 W8BRW9 R4WR60 A0A1L8EGK8 W8BED8 A0A0K8W9F6 A0A1I8PH54 A0A0Q9WP14 A0A034VDF6 A0A0K8UKJ5 A0A1I8NAH3 B4QB34 A0A0B4KEL2 A0A034V927 W8C1R3 A0A0N7ZXB1 A0A336LV26 E9GIA0 A0A0P5HUT3 W5J5J0 A0A0P5T9E0 A0A0P5A6Q2 A0A1Y1JWZ4 A0A067QG92 C3ZIU3 A0A182YJI1 A0A182SMD3 A0A1W4WF47 A0A1W4WQ33 A0A1B6HDV9 A0A1B6LRE2 A0A0P6CRD8 A0A0P6CLZ2 A0A182L446 A0A0P5SL04 A0A182R9A9 A0A060YVX0 A0A1L8E3L1 A0A1B6LTY2 T1IB30 A0A182X243 A0A182N7I8 A0A182I6A4 E0VXM0 A0A1S3KKV9 A0A0A9XAE0 A0A182VGT9 A0A2J7R040 A0A182QYG9 A0A2J7R027 A0A2D0R9Y6 A0A2J7R038 A0A1B6E985
A0A0N0PAM0 A0A1B6J2B1 A0A1B6DVY3 A0A2J7QX24 A0A2J7QX19 V5GLD7 V5GX66 A0A1Y1NF14 A0A139WNT6 D6W758 A0A023FBA2 A0A067RC55 A0A224XHI0 A0A146LE77 A0A146KLW3 A0A2P8Y3W2 J3JUV8 N6T2A2 B3MEQ8 B4P7K7 A0A023FB97 B4J846 B3NSN2 B4MK84 A0A1W4U5M6 B4KQ27 Q28WS6 B4H6B7 A1Z8F5 B4ME12 A0A3B0JL62 Q8SZ85 B5RJ61 A0A0A1XPE8 A0A0Q9WBG8 A0A0L0BP84 A0A3B0J034 B4HN40 W8BRW9 R4WR60 A0A1L8EGK8 W8BED8 A0A0K8W9F6 A0A1I8PH54 A0A0Q9WP14 A0A034VDF6 A0A0K8UKJ5 A0A1I8NAH3 B4QB34 A0A0B4KEL2 A0A034V927 W8C1R3 A0A0N7ZXB1 A0A336LV26 E9GIA0 A0A0P5HUT3 W5J5J0 A0A0P5T9E0 A0A0P5A6Q2 A0A1Y1JWZ4 A0A067QG92 C3ZIU3 A0A182YJI1 A0A182SMD3 A0A1W4WF47 A0A1W4WQ33 A0A1B6HDV9 A0A1B6LRE2 A0A0P6CRD8 A0A0P6CLZ2 A0A182L446 A0A0P5SL04 A0A182R9A9 A0A060YVX0 A0A1L8E3L1 A0A1B6LTY2 T1IB30 A0A182X243 A0A182N7I8 A0A182I6A4 E0VXM0 A0A1S3KKV9 A0A0A9XAE0 A0A182VGT9 A0A2J7R040 A0A182QYG9 A0A2J7R027 A0A2D0R9Y6 A0A2J7R038 A0A1B6E985
Ontologies
PANTHER
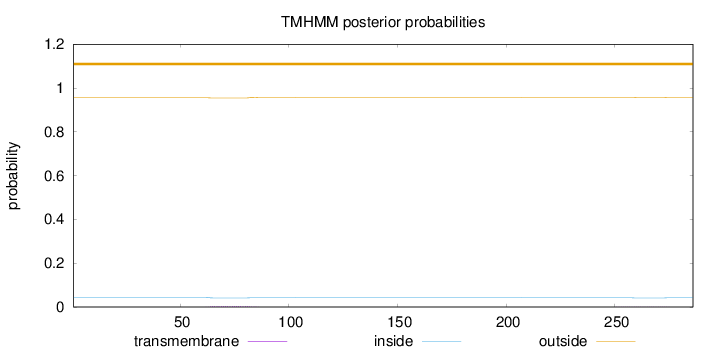
Topology
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0943600000000001
Exp number, first 60 AAs:
6e-05
Total prob of N-in:
0.04469
outside
1 - 286
Population Genetic Test Statistics
Pi
279.973367
Theta
171.311639
Tajima's D
1.844597
CLR
0.499804
CSRT
0.854357282135893
Interpretation
Uncertain
