Gene
KWMTBOMO02995
Pre Gene Modal
BGIBMGA003617
Annotation
PREDICTED:_ankyrin_repeat_and_MYND_domain-containing_protein_1-like_[Amyelois_transitella]
Full name
Ankyrin repeat and MYND domain-containing protein 1
Location in the cell
Nuclear Reliability : 3.075
Sequence
CDS
ATGCACGGAATGGGCGAATATAAATGGCAGCATCAAGGTCCGAATGATATATTTGCTAGCTACGAAGGATATTTTTACGCCAATACAATGCATGGTTACGGCACTATGTCATATGCAGACGGAAGAGTTTTTAGTGGATTATATTATAACAATATGCGTTGGGGGCCAGGTATTGAATCCCATATTAATTTACAAGAAAACGTGGGATTGTGGCGGGGGAGTCAGTTGATTCGTTTGGCCTGGCGACCACAAGCACCAAGCATAACCCTGGACCTGACCTCGGCTGCTAATGGAAAAGATTTCGTTGAATCGTATCGCACTATGCTCCTACCAAAACCAAGTACAGTTGATGAAATAAATAGTGCTGTAGACTTATTGAAGGATTCCGGGTGTGACCCGTTACTGGCCTCAAAAATATGGCATAAATTATATCCTAAAAATTGCACCAACGTGAAAAGCGCTGCTTGTAATATTCAACTGTTTGAACGTAATTACTATACTGAAGGCTGTTACTCTTTAGAAGAATTACACGAATGGAATGATGAGATAACAGACGCAGAACCAAATCTAGAAATCCATTACGCTTGGAATAATAGTAAAACAATTTTGCACATTATGAAGCATTGTTATAGACATGAAAATCAAAGATCGACTTATAACATAAATATAGCGGGCATACTAAGTGGACAAAGATTTTTGTTTAAATCTCCCGGAATACACGAATTAAATTGCAGGACGCTTCTGATGGCCTCTTATCTCGGATACCTTTCCAAAGTAGCGCAACTTATAATGGAGAGCAACGTGCATCCAGATGTAGCAGACTGCCAAGGGAATTCAACCATTATGTACGCAACTGCGGGTGAACAAGTTGAGATAATTCTATTTTTGGTGGAGGTTGGCGCTGACATTAATTGTTATAATGACTCATGTTGTACGCCGCTTGGTTTAGCTTTGATGAAACTGCTATGTGTCCAAAATGATATACCCATATCGACAATAGTTCAGTCTTTATTACCACCAACTTCTACTCAACAGTCATGTACAGATAAAATAGAAAATGTGTCTGAATGGAATATTGTGAGAGAGACACCTGCTCTCAGCCCTGGTCGACCTTTTACAAGATCTCCAAGTAAAGTCAAAGCACCAAACACGAAAAAGCTAAAGTCTTCAATTTCACTAAAGGACGCACCGAAAAGGAAACAAGAATTATTGTTTGAAAAAGAAATAAACTCCGACACTAACTCTTTTGAGTCATTTTCTGATAGCGAAAGACTTTATACCAGTATAAACAGTGAATTTATAATGAAGATCGATGATTTATTCCAACAAACAACAGCCAACAATATTCTGTTCTATCTTTTCGAAGTGCACGATATGGTCAAAGAAGTAATTGAAAACGATGAAGATCATAAAAAAGCGACCGAGAAAAATCCAAAGAAAGCATTATCTAAGGAATATAAAGTCAAAACGAAATTAAGTAAAGAATTGTTTCCACAAATTGTTGAAGATTCTTCATCTATCACTAGCGCGGATAAAGCTAAAAAGGAGACTTTAGATAGAATTAAGCTTACGATACTCCAGTTATGTGATAATGGTGCGGATCCTTGCTTAGTGAACTGTCCACAGCCATCCCTGGTAATGGCGATACTCGCTAGTTGTCCGGAAATAATTTCTGAGCTTGTAAGCCGAGGAGCTGATATAAATATGAACTATTCCAATTTTAGAAATTACACCGCTTTAGACTTGGCAATTTCACTTCCATTTACATTTGTGAATTTAGAAACGATTCGTACCATTCTAAAAGCCGGCGGAAAGGCAAACCATTTACTGCAAAGTATGGAGTATCCAGAATACGATAATAATGGACCGACATTGTTACACGTTGTTCTTTCAAAGACAACAGAAAGTGATACTGAGGAAGAGATACGTGGTCATATTATAGCGCTCCTTCTAGAGCATGACTGTGATCCAACCATTCAGTTCAAAGGGAGGTCACCTATGGACATTGCCCTTTCAAAGAGTTTGGATATTTTTGATATATTTGTAAAATCACCGAAGACCAATCTTAATACTGTTATTAATAATAGCAATCAAAATGTTCTAGTGAAGTTGTTTGTCGCAAATTTTTGTAATACTTTATCATTGACGGATAGATTACAGATCTTAACGAATTTAATGCAACGCGGCGCGAATCCTTTGCTGGAATGTTACAATGGCTCTGAAAAATATCAGAATTTATTTGTTTTTGCAAATAAAATTATGGAAAATTTTGAATCGCAGCCGAAACAGCACGATACAAAACCGAAGAAAGCTGATAAACCCAAGAAAGAGGACAAGCTAGCTAGTAAATCTTTAGGAAAATTAACCTCTGACGAAGTTGAAGATTACAAACAAGCACTACAGTTGGTTAAGGAATACGCCAGATATTTACATATCACGTGGATACAGATAAAACTGGTTAAGGAATTGTCAGACATAGTCCAAAGATATAAACACCGACACTGGAGCATGATCATTAAGGAACATAAAAATTGGAAACAAATGAAATTATGGGTAACAACACGAAGATGTTTGGAGATATGGAACGTTTTAAGAAGTAGTCAGAAATTTGTAAATGGGAAGAATATTTTAAAAAACCTTTTGTGCATTGTTGAATTTTATAATGTTACGCCATTTGGAAAATCATTAACAAAAATTACTGCAGAATTAAAAGACAGCGTAGAAAAACAAGTTAAAGGAATTCTACGTGAACAAGAACTTAGTAAAGCTATGTCAGATTCCACGTTGTCTTATGTTGCATTTGAGCTGAAGGAAGATGCAGTTAGTTTTTTTTTTTAG
Protein
MHGMGEYKWQHQGPNDIFASYEGYFYANTMHGYGTMSYADGRVFSGLYYNNMRWGPGIESHINLQENVGLWRGSQLIRLAWRPQAPSITLDLTSAANGKDFVESYRTMLLPKPSTVDEINSAVDLLKDSGCDPLLASKIWHKLYPKNCTNVKSAACNIQLFERNYYTEGCYSLEELHEWNDEITDAEPNLEIHYAWNNSKTILHIMKHCYRHENQRSTYNINIAGILSGQRFLFKSPGIHELNCRTLLMASYLGYLSKVAQLIMESNVHPDVADCQGNSTIMYATAGEQVEIILFLVEVGADINCYNDSCCTPLGLALMKLLCVQNDIPISTIVQSLLPPTSTQQSCTDKIENVSEWNIVRETPALSPGRPFTRSPSKVKAPNTKKLKSSISLKDAPKRKQELLFEKEINSDTNSFESFSDSERLYTSINSEFIMKIDDLFQQTTANNILFYLFEVHDMVKEVIENDEDHKKATEKNPKKALSKEYKVKTKLSKELFPQIVEDSSSITSADKAKKETLDRIKLTILQLCDNGADPCLVNCPQPSLVMAILASCPEIISELVSRGADINMNYSNFRNYTALDLAISLPFTFVNLETIRTILKAGGKANHLLQSMEYPEYDNNGPTLLHVVLSKTTESDTEEEIRGHIIALLLEHDCDPTIQFKGRSPMDIALSKSLDIFDIFVKSPKTNLNTVINNSNQNVLVKLFVANFCNTLSLTDRLQILTNLMQRGANPLLECYNGSEKYQNLFVFANKIMENFESQPKQHDTKPKKADKPKKEDKLASKSLGKLTSDEVEDYKQALQLVKEYARYLHITWIQIKLVKELSDIVQRYKHRHWSMIIKEHKNWKQMKLWVTTRRCLEIWNVLRSSQKFVNGKNILKNLLCIVEFYNVTPFGKSLTKITAELKDSVEKQVKGILREQELSKAMSDSTLSYVAFELKEDAVSFFF
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Keywords
ANK repeat
Complete proteome
Metal-binding
Reference proteome
Repeat
Zinc
Zinc-finger
Feature
chain Ankyrin repeat and MYND domain-containing protein 1
Uniprot
A0A212EVG1
A0A2H1W974
A0A194PNU0
A0A0L7LP82
A0A0N0PAM0
A0A2A4JFP6
+ More
A0A2C9JZC2 A0A2C9JZE1 A0A2C9JZC5 A0A1S3K7W5 A0A3L7IEJ8 A0A2T7ND73 G3H4T6 V4A115 R7U1E8 G1KAI1 A0A2I2UVL7 E0CYY3 E9PUG3 Q8C0W1 A0A1W4XQY8 A0A0P7ULC1 A0A1A6HSK5 A0A1U7R113 M7B0M0 A0A3B4CV31 A0A3B4EE17 A0A151NUA1 H0XES3 F1PP72 A0A2Y9IRA6 H2ZVI2 A0A1U7SX57 G1MEP8 A0A2Y9GA85 D2HLE3 A0A384C0Y5 K7FQI8
A0A2C9JZC2 A0A2C9JZE1 A0A2C9JZC5 A0A1S3K7W5 A0A3L7IEJ8 A0A2T7ND73 G3H4T6 V4A115 R7U1E8 G1KAI1 A0A2I2UVL7 E0CYY3 E9PUG3 Q8C0W1 A0A1W4XQY8 A0A0P7ULC1 A0A1A6HSK5 A0A1U7R113 M7B0M0 A0A3B4CV31 A0A3B4EE17 A0A151NUA1 H0XES3 F1PP72 A0A2Y9IRA6 H2ZVI2 A0A1U7SX57 G1MEP8 A0A2Y9GA85 D2HLE3 A0A384C0Y5 K7FQI8
Pubmed
EMBL
AGBW02012195
OWR45473.1
ODYU01007111
SOQ49597.1
KQ459597
KPI95096.1
+ More
JTDY01000442 KOB77154.1 KQ461187 KPJ07340.1 NWSH01001547 PCG70897.1 RAZU01000074 RLQ75762.1 PZQS01000014 PVD19092.1 JH000147 EGV95317.1 KB202656 ESO88630.1 AMQN01002176 KB308559 ELT97476.1 AANG04002752 AC131059 CH466520 EDL39990.1 AK029700 BC120887 BC120888 JARO02008553 KPP62579.1 LZPO01017287 OBS81204.1 KB545796 EMP31371.1 AKHW03001930 KYO40437.1 AAQR03144419 AAQR03144420 AAQR03144421 AAQR03144422 AAQR03144423 AAEX03014534 AFYH01246016 AFYH01246017 AFYH01246018 AFYH01246019 AFYH01246020 AFYH01246021 AFYH01246022 ACTA01003225 ACTA01011225 ACTA01019225 ACTA01027225 GL193002 EFB16591.1 AGCU01100967 AGCU01100968 AGCU01100969 AGCU01100970 AGCU01100971 AGCU01100972 AGCU01100973 AGCU01100974 AGCU01100975
JTDY01000442 KOB77154.1 KQ461187 KPJ07340.1 NWSH01001547 PCG70897.1 RAZU01000074 RLQ75762.1 PZQS01000014 PVD19092.1 JH000147 EGV95317.1 KB202656 ESO88630.1 AMQN01002176 KB308559 ELT97476.1 AANG04002752 AC131059 CH466520 EDL39990.1 AK029700 BC120887 BC120888 JARO02008553 KPP62579.1 LZPO01017287 OBS81204.1 KB545796 EMP31371.1 AKHW03001930 KYO40437.1 AAQR03144419 AAQR03144420 AAQR03144421 AAQR03144422 AAQR03144423 AAEX03014534 AFYH01246016 AFYH01246017 AFYH01246018 AFYH01246019 AFYH01246020 AFYH01246021 AFYH01246022 ACTA01003225 ACTA01011225 ACTA01019225 ACTA01027225 GL193002 EFB16591.1 AGCU01100967 AGCU01100968 AGCU01100969 AGCU01100970 AGCU01100971 AGCU01100972 AGCU01100973 AGCU01100974 AGCU01100975
Proteomes
UP000007151
UP000053268
UP000037510
UP000053240
UP000218220
UP000076420
+ More
UP000085678 UP000273346 UP000245119 UP000001075 UP000030746 UP000014760 UP000001646 UP000011712 UP000000589 UP000192224 UP000034805 UP000092124 UP000189706 UP000031443 UP000261440 UP000050525 UP000005225 UP000002254 UP000248482 UP000008672 UP000189704 UP000008912 UP000248481 UP000261680 UP000007267
UP000085678 UP000273346 UP000245119 UP000001075 UP000030746 UP000014760 UP000001646 UP000011712 UP000000589 UP000192224 UP000034805 UP000092124 UP000189706 UP000031443 UP000261440 UP000050525 UP000005225 UP000002254 UP000248482 UP000008672 UP000189704 UP000008912 UP000248481 UP000261680 UP000007267
Pfam
Interpro
IPR002110
Ankyrin_rpt
+ More
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR003409 MORN
IPR005552 Scramblase
IPR002893 Znf_MYND
IPR032405 Kinesin_assoc
IPR036961 Kinesin_motor_dom_sf
IPR011993 PH-like_dom_sf
IPR022140 Kinesin-like_KIF1-typ
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR001849 PH_domain
IPR000253 FHA_dom
IPR022164 Kinesin-like
IPR008984 SMAD_FHA_dom_sf
IPR017974 Claudin_CS
IPR004031 PMP22/EMP/MP20/Claudin
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR003409 MORN
IPR005552 Scramblase
IPR002893 Znf_MYND
IPR032405 Kinesin_assoc
IPR036961 Kinesin_motor_dom_sf
IPR011993 PH-like_dom_sf
IPR022140 Kinesin-like_KIF1-typ
IPR027640 Kinesin-like_fam
IPR001752 Kinesin_motor_dom
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR001849 PH_domain
IPR000253 FHA_dom
IPR022164 Kinesin-like
IPR008984 SMAD_FHA_dom_sf
IPR017974 Claudin_CS
IPR004031 PMP22/EMP/MP20/Claudin
Gene 3D
ProteinModelPortal
A0A212EVG1
A0A2H1W974
A0A194PNU0
A0A0L7LP82
A0A0N0PAM0
A0A2A4JFP6
+ More
A0A2C9JZC2 A0A2C9JZE1 A0A2C9JZC5 A0A1S3K7W5 A0A3L7IEJ8 A0A2T7ND73 G3H4T6 V4A115 R7U1E8 G1KAI1 A0A2I2UVL7 E0CYY3 E9PUG3 Q8C0W1 A0A1W4XQY8 A0A0P7ULC1 A0A1A6HSK5 A0A1U7R113 M7B0M0 A0A3B4CV31 A0A3B4EE17 A0A151NUA1 H0XES3 F1PP72 A0A2Y9IRA6 H2ZVI2 A0A1U7SX57 G1MEP8 A0A2Y9GA85 D2HLE3 A0A384C0Y5 K7FQI8
A0A2C9JZC2 A0A2C9JZE1 A0A2C9JZC5 A0A1S3K7W5 A0A3L7IEJ8 A0A2T7ND73 G3H4T6 V4A115 R7U1E8 G1KAI1 A0A2I2UVL7 E0CYY3 E9PUG3 Q8C0W1 A0A1W4XQY8 A0A0P7ULC1 A0A1A6HSK5 A0A1U7R113 M7B0M0 A0A3B4CV31 A0A3B4EE17 A0A151NUA1 H0XES3 F1PP72 A0A2Y9IRA6 H2ZVI2 A0A1U7SX57 G1MEP8 A0A2Y9GA85 D2HLE3 A0A384C0Y5 K7FQI8
PDB
5GIK
E-value=0.00177541,
Score=102
Ontologies
GO
PANTHER
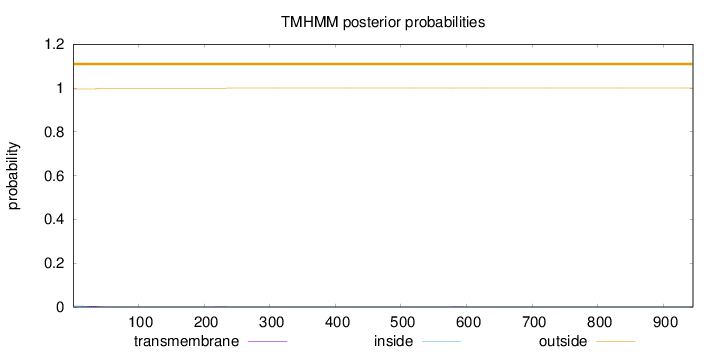
Topology
Length:
945
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0953000000000001
Exp number, first 60 AAs:
0.08321
Total prob of N-in:
0.00430
outside
1 - 945
Population Genetic Test Statistics
Pi
170.271517
Theta
153.387535
Tajima's D
0.381357
CLR
179.490896
CSRT
0.47817609119544
Interpretation
Uncertain
