Gene
KWMTBOMO02994
Pre Gene Modal
BGIBMGA003618
Annotation
PREDICTED:_deoxyribodipyrimidine_photo-lyase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.285
Sequence
CDS
ATGCTTCATATTCCACTTGTAAATCAATTTAATACGGTTTTAAAACGCTATTCTGTATTCATAATGGCATCGGCTTCAAAGAAACGCAAAACTTCCATTCCTATGGCATTAGGTAAAAGCAATGAAAGTACAAGCACCATTGAAGAGTTTCTGAAAAAACTACAAAAAAAGCGCGAAGAAACGGCTGAGTCTATTTTGAACTTTGATTTCAACAAAAAACGCATTCGAATAATATCTCAAGAACAAATTATACCTGACGATTGCGATGGTGTTGTATACTGGATGTCTAGAGACAGCCGTGTTCAGGATAATTGGGCTTTCCTTTTTGCACAAAAACTGGCCTTGAAAAACGAAGTACCGCTTCATGTATGTTTTTGTTTAATTTCGAAGTATCTGGATGCGTCCTTGCGACAATTTCATTTCCTTGTAAAAGGACTAGAAAAAGTAGCTGCAGAGTGTAAGAAACTAAACATACAATTCCATTTATTAGAAGGTAGTGGTGCGGAAGTGTTACCTCAATGGGTAGTAGATCATAATATTGGTGCGGTTGTGTGTGATTTCAATCCACTTAGGGTCCCTATGGGCTGGTTGGATGGAGTAAAGGGAAAACTTAGGAAAGATGTGCCTTTAATTCAGGTAGACACTCACAATGTAGTTCCATGTTGGGTGGCATCAGACAAACAGGAGTATTCTGCAAGGACAATCAGGAATAAAATTAATTCAAAACTTGATGAGTTTCTCACAGAATTCCCCCCAGTTATAAAACATCCATATTCCAGCAAATTTGAAGCAGAGCCCATTGACTGGCATGAAGCCATTGAGACGAGAGAAGCAGATAAATCAGTTGGTCCTGTTGATTGGGCTAAGCCTGGTTATGATGAAGCTGTGAAAATGATGAAAAGTTTTCTTGACAAGAGATTAAACATATATGCCAGTAAAAGGAATGACCCAACTCAGGATGCCCTGAGCAATCTCTCACCTTGGTTCCACTTTGGTCAAATATCAGTACAAAGGGTAGCCCTATGTGTACAAGAATACAAATCGAAATTTACTGAAAGTGTGAATGCATTTCTAGAAGAAGCTATAGTAAGGCGAGAACTAGCTGACAACTTTTGTTTCTATTGTGAACATTACGACAGTGTAAAGGGTGCTAGTGCGTGGGCCCAAAAAACTTTGGATGATCACAGAAAAGACAAGAGAACTCACATTTATACTCTAGAAAAACTTTCTAAAGGTGAAACTCATGATGACTTGTGGAATTCGGCTCAGCTACAACTTGTTAAAGAGGGCAAAATGCATGGCTTTTTAAGAATGTATTGGTGTAAGAAAATATTGGAATGGACTCCTACTCCGGAGGATGCTCTTAAATATGGAATATATTTGAACGACCACTATAGTATCGATGGGAGAGATCCAAGTGGATTTGTAGGCTGCATGTGGTCGATATGCGGCATCCACGACCAAGGCTGGGCAGAGCGCGCCGTCTTCGGTAAAATTAGATACATGAATTACGACGGGTGTAAGCGTAAGTTCGACATCAAAGCCTTCATTGCCAGGTACGGAGGGAAGGTACACAAATATGTACCTAAAAAGTAG
Protein
MLHIPLVNQFNTVLKRYSVFIMASASKKRKTSIPMALGKSNESTSTIEEFLKKLQKKREETAESILNFDFNKKRIRIISQEQIIPDDCDGVVYWMSRDSRVQDNWAFLFAQKLALKNEVPLHVCFCLISKYLDASLRQFHFLVKGLEKVAAECKKLNIQFHLLEGSGAEVLPQWVVDHNIGAVVCDFNPLRVPMGWLDGVKGKLRKDVPLIQVDTHNVVPCWVASDKQEYSARTIRNKINSKLDEFLTEFPPVIKHPYSSKFEAEPIDWHEAIETREADKSVGPVDWAKPGYDEAVKMMKSFLDKRLNIYASKRNDPTQDALSNLSPWFHFGQISVQRVALCVQEYKSKFTESVNAFLEEAIVRRELADNFCFYCEHYDSVKGASAWAQKTLDDHRKDKRTHIYTLEKLSKGETHDDLWNSAQLQLVKEGKMHGFLRMYWCKKILEWTPTPEDALKYGIYLNDHYSIDGRDPSGFVGCMWSICGIHDQGWAERAVFGKIRYMNYDGCKRKFDIKAFIARYGGKVHKYVPKK
Summary
Uniprot
H9J281
A0A2A4JGI9
A0A0N1I4S2
A0A194PQ52
A0A194PVE0
A0A0N1PGJ5
+ More
A0A182NZR1 A0A182H124 A0A182HPD9 Q17KA3 A0A084WCU3 A0A182TDL1 A0A182V452 A0A182X0X1 A0A182LB47 B0WAC3 A0A182WMB2 A0A1Q3F590 A0A182FY44 Q7Q9T5 A0A1B6H369 W5J446 A0A182NLW6 A0A182LSK5 A0A182RUX5 A0A182QNV4 A0A182YJ19 A0A1B6KN13 F6W9G4 A0A1B6EB20 A0A336L2S2 Q3C2L7 A0A336LWB1 A0A182JL00 A0A1J1J7Y8 A0A0L0BRI0 A0A1S3H867 A0A0A9WTY5 A0A2S2QTI6 V4A167 B3MG96 A0A0P8Y6L2 A0A0P4WEL0 A0A075F4M7 B4KQ29 A0A2S2NFH5 A0A2I0U149 A0A210PRW9 Q24443 Q28ZN6 J9K3D6 A0A0N1INB4 T1L328 B4GH10 B4QET3 A0A0A1XCX0 A1Z757 B4HR84 Q7JY97 K7J6K2 J3S999 A0A1W4VZF9 T1HKC6 A0A3B0J6V3 A0A1I8MLY4 A0A0K8RWA2 B3N9D8 A0A2A3EEH9 K1PPR1 A0A1I8MLY6 Q26373 B4J848 A0A0B7AUQ3 A0A232EFC2 A0A0K8V113 B4ME10 A0A1V4KMC1 W8C169 A0A088ARG8 A0A2B4RXJ6 B4P244 A0A0C9Q5W8 H9G6X4 A0A023F3G0 A0A194PQS4 A0A034V6B8 F6Q9E0 A0A0M4E6S9 A0A034V448 A0A1V4KMH6 A0A2I0LMR4 Q28464 A0A3M7QL57 B4MQY9 E9HAM9 G3VYP1 S4R979 A0A0P6JIK5 A0A151MZN9 A0A0N8CYE9
A0A182NZR1 A0A182H124 A0A182HPD9 Q17KA3 A0A084WCU3 A0A182TDL1 A0A182V452 A0A182X0X1 A0A182LB47 B0WAC3 A0A182WMB2 A0A1Q3F590 A0A182FY44 Q7Q9T5 A0A1B6H369 W5J446 A0A182NLW6 A0A182LSK5 A0A182RUX5 A0A182QNV4 A0A182YJ19 A0A1B6KN13 F6W9G4 A0A1B6EB20 A0A336L2S2 Q3C2L7 A0A336LWB1 A0A182JL00 A0A1J1J7Y8 A0A0L0BRI0 A0A1S3H867 A0A0A9WTY5 A0A2S2QTI6 V4A167 B3MG96 A0A0P8Y6L2 A0A0P4WEL0 A0A075F4M7 B4KQ29 A0A2S2NFH5 A0A2I0U149 A0A210PRW9 Q24443 Q28ZN6 J9K3D6 A0A0N1INB4 T1L328 B4GH10 B4QET3 A0A0A1XCX0 A1Z757 B4HR84 Q7JY97 K7J6K2 J3S999 A0A1W4VZF9 T1HKC6 A0A3B0J6V3 A0A1I8MLY4 A0A0K8RWA2 B3N9D8 A0A2A3EEH9 K1PPR1 A0A1I8MLY6 Q26373 B4J848 A0A0B7AUQ3 A0A232EFC2 A0A0K8V113 B4ME10 A0A1V4KMC1 W8C169 A0A088ARG8 A0A2B4RXJ6 B4P244 A0A0C9Q5W8 H9G6X4 A0A023F3G0 A0A194PQS4 A0A034V6B8 F6Q9E0 A0A0M4E6S9 A0A034V448 A0A1V4KMH6 A0A2I0LMR4 Q28464 A0A3M7QL57 B4MQY9 E9HAM9 G3VYP1 S4R979 A0A0P6JIK5 A0A151MZN9 A0A0N8CYE9
Pubmed
19121390
26354079
26483478
17510324
24438588
20966253
+ More
12364791 14747013 17210077 20920257 23761445 25244985 20431018 16302973 26108605 26383154 25401762 23254933 17994087 24568948 28812685 7813451 15632085 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 23025625 25315136 25727380 22992520 7526199 28648823 24495485 17550304 21881562 25474469 25348373 17495919 23371554 7937136 30375419 21292972 21709235 22293439
12364791 14747013 17210077 20920257 23761445 25244985 20431018 16302973 26108605 26383154 25401762 23254933 17994087 24568948 28812685 7813451 15632085 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 23025625 25315136 25727380 22992520 7526199 28648823 24495485 17550304 21881562 25474469 25348373 17495919 23371554 7937136 30375419 21292972 21709235 22293439
EMBL
BABH01007436
NWSH01001547
PCG70896.1
KQ461187
KPJ07337.1
KQ459597
+ More
KPI95093.1 KPI95095.1 KPJ07339.1 JXUM01102357 KQ564721 KXJ71848.1 APCN01002982 CH477227 EAT47082.1 ATLV01022885 KE525337 KFB48037.1 DS231870 EDS41013.1 GFDL01012315 JAV22730.1 AAAB01008900 EAA09350.3 GECZ01000640 JAS69129.1 ADMH02002165 ETN58118.1 AXCM01004675 AXCN02000947 GEBQ01027138 JAT12839.1 AAMC01069403 AAMC01069404 AAMC01069405 AAMC01069406 AAMC01069407 GEDC01009216 GEDC01002224 JAS28082.1 JAS35074.1 UFQS01001398 UFQT01001398 SSX10693.1 SSX30375.1 BC133744 AB175838 AAI33745.1 BAE46749.1 UFQT01000246 SSX22346.1 CVRI01000075 CRL08549.1 JRES01001578 KNC21824.1 GBHO01032723 JAG10881.1 GGMS01011852 MBY81055.1 KB202656 ESO88680.1 CH902619 EDV37799.1 KPU77038.1 GDRN01063567 JAI64957.1 KJ438858 AIE57497.1 CH933808 EDW08131.2 GGMR01003249 MBY15868.1 KZ506404 PKU39806.1 NEDP02005537 OWF39196.1 D26021 BAA05042.1 CM000071 EAL25577.4 ABLF02030880 KPJ07338.1 CAEY01001007 CH479183 EDW35780.1 CM000362 EDX06079.1 GBXI01005989 JAD08303.1 AE013599 AAM68868.1 CH480816 EDW46827.1 AY084133 AAF59185.1 AAL89871.1 AAZX01001310 JU175419 AFJ50943.1 ACPB03011894 OUUW01000001 SPP75572.1 GBKC01000971 JAG45099.1 CH954177 EDV59625.2 KZ288282 PBC29576.1 JH823231 EKC26192.1 S73530 AAB32328.1 CH916367 EDW01185.1 HACG01037698 CEK84563.1 NNAY01005071 OXU17043.1 GDHF01019768 JAI32546.1 CH940662 EDW58775.2 LSYS01002834 OPJ85594.1 GAMC01003687 JAC02869.1 LSMT01000243 PFX22341.1 CM000157 EDW89245.2 GBYB01009508 JAG79275.1 AAWZ02027807 AAWZ02027808 GBBI01002906 JAC15806.1 KPI95094.1 GAKP01020893 JAC38059.1 CP012524 ALC42266.1 GAKP01020898 GAKP01020896 GAKP01020895 JAC38056.1 OPJ85593.1 AKCR02000185 PKK18718.1 D31902 BAA06700.1 REGN01005786 RNA12022.1 CH963849 EDW74528.1 GL732612 EFX71237.1 AEFK01107965 AEFK01107966 AEFK01107967 GDIQ01006883 JAN87854.1 AKHW03004458 KYO29905.1 GDIP01086655 JAM17060.1
KPI95093.1 KPI95095.1 KPJ07339.1 JXUM01102357 KQ564721 KXJ71848.1 APCN01002982 CH477227 EAT47082.1 ATLV01022885 KE525337 KFB48037.1 DS231870 EDS41013.1 GFDL01012315 JAV22730.1 AAAB01008900 EAA09350.3 GECZ01000640 JAS69129.1 ADMH02002165 ETN58118.1 AXCM01004675 AXCN02000947 GEBQ01027138 JAT12839.1 AAMC01069403 AAMC01069404 AAMC01069405 AAMC01069406 AAMC01069407 GEDC01009216 GEDC01002224 JAS28082.1 JAS35074.1 UFQS01001398 UFQT01001398 SSX10693.1 SSX30375.1 BC133744 AB175838 AAI33745.1 BAE46749.1 UFQT01000246 SSX22346.1 CVRI01000075 CRL08549.1 JRES01001578 KNC21824.1 GBHO01032723 JAG10881.1 GGMS01011852 MBY81055.1 KB202656 ESO88680.1 CH902619 EDV37799.1 KPU77038.1 GDRN01063567 JAI64957.1 KJ438858 AIE57497.1 CH933808 EDW08131.2 GGMR01003249 MBY15868.1 KZ506404 PKU39806.1 NEDP02005537 OWF39196.1 D26021 BAA05042.1 CM000071 EAL25577.4 ABLF02030880 KPJ07338.1 CAEY01001007 CH479183 EDW35780.1 CM000362 EDX06079.1 GBXI01005989 JAD08303.1 AE013599 AAM68868.1 CH480816 EDW46827.1 AY084133 AAF59185.1 AAL89871.1 AAZX01001310 JU175419 AFJ50943.1 ACPB03011894 OUUW01000001 SPP75572.1 GBKC01000971 JAG45099.1 CH954177 EDV59625.2 KZ288282 PBC29576.1 JH823231 EKC26192.1 S73530 AAB32328.1 CH916367 EDW01185.1 HACG01037698 CEK84563.1 NNAY01005071 OXU17043.1 GDHF01019768 JAI32546.1 CH940662 EDW58775.2 LSYS01002834 OPJ85594.1 GAMC01003687 JAC02869.1 LSMT01000243 PFX22341.1 CM000157 EDW89245.2 GBYB01009508 JAG79275.1 AAWZ02027807 AAWZ02027808 GBBI01002906 JAC15806.1 KPI95094.1 GAKP01020893 JAC38059.1 CP012524 ALC42266.1 GAKP01020898 GAKP01020896 GAKP01020895 JAC38056.1 OPJ85593.1 AKCR02000185 PKK18718.1 D31902 BAA06700.1 REGN01005786 RNA12022.1 CH963849 EDW74528.1 GL732612 EFX71237.1 AEFK01107965 AEFK01107966 AEFK01107967 GDIQ01006883 JAN87854.1 AKHW03004458 KYO29905.1 GDIP01086655 JAM17060.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000075885
UP000069940
+ More
UP000249989 UP000075840 UP000008820 UP000030765 UP000075902 UP000075903 UP000076407 UP000075882 UP000002320 UP000075920 UP000069272 UP000007062 UP000000673 UP000075884 UP000075883 UP000075900 UP000075886 UP000076408 UP000008143 UP000075880 UP000183832 UP000037069 UP000085678 UP000030746 UP000007801 UP000009192 UP000242188 UP000001819 UP000007819 UP000015104 UP000008744 UP000000304 UP000000803 UP000001292 UP000002358 UP000192221 UP000015103 UP000268350 UP000095301 UP000008711 UP000242457 UP000005408 UP000001070 UP000215335 UP000008792 UP000190648 UP000005203 UP000225706 UP000002282 UP000001646 UP000002280 UP000092553 UP000053872 UP000276133 UP000007798 UP000000305 UP000007648 UP000245300 UP000050525
UP000249989 UP000075840 UP000008820 UP000030765 UP000075902 UP000075903 UP000076407 UP000075882 UP000002320 UP000075920 UP000069272 UP000007062 UP000000673 UP000075884 UP000075883 UP000075900 UP000075886 UP000076408 UP000008143 UP000075880 UP000183832 UP000037069 UP000085678 UP000030746 UP000007801 UP000009192 UP000242188 UP000001819 UP000007819 UP000015104 UP000008744 UP000000304 UP000000803 UP000001292 UP000002358 UP000192221 UP000015103 UP000268350 UP000095301 UP000008711 UP000242457 UP000005408 UP000001070 UP000215335 UP000008792 UP000190648 UP000005203 UP000225706 UP000002282 UP000001646 UP000002280 UP000092553 UP000053872 UP000276133 UP000007798 UP000000305 UP000007648 UP000245300 UP000050525
Interpro
IPR006050
DNA_photolyase_N
+ More
IPR032673 DNA_photolyase_2_CS
IPR036134 Crypto/Photolyase_FAD-like_sf
IPR014729 Rossmann-like_a/b/a_fold
IPR036155 Crypto/Photolyase_N_sf
IPR008148 DNA_photolyase_2
IPR037431 REX4_DEDDh_dom
IPR012337 RNaseH-like_sf
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR036397 RNaseH_sf
IPR032673 DNA_photolyase_2_CS
IPR036134 Crypto/Photolyase_FAD-like_sf
IPR014729 Rossmann-like_a/b/a_fold
IPR036155 Crypto/Photolyase_N_sf
IPR008148 DNA_photolyase_2
IPR037431 REX4_DEDDh_dom
IPR012337 RNaseH-like_sf
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR036397 RNaseH_sf
Gene 3D
ProteinModelPortal
H9J281
A0A2A4JGI9
A0A0N1I4S2
A0A194PQ52
A0A194PVE0
A0A0N1PGJ5
+ More
A0A182NZR1 A0A182H124 A0A182HPD9 Q17KA3 A0A084WCU3 A0A182TDL1 A0A182V452 A0A182X0X1 A0A182LB47 B0WAC3 A0A182WMB2 A0A1Q3F590 A0A182FY44 Q7Q9T5 A0A1B6H369 W5J446 A0A182NLW6 A0A182LSK5 A0A182RUX5 A0A182QNV4 A0A182YJ19 A0A1B6KN13 F6W9G4 A0A1B6EB20 A0A336L2S2 Q3C2L7 A0A336LWB1 A0A182JL00 A0A1J1J7Y8 A0A0L0BRI0 A0A1S3H867 A0A0A9WTY5 A0A2S2QTI6 V4A167 B3MG96 A0A0P8Y6L2 A0A0P4WEL0 A0A075F4M7 B4KQ29 A0A2S2NFH5 A0A2I0U149 A0A210PRW9 Q24443 Q28ZN6 J9K3D6 A0A0N1INB4 T1L328 B4GH10 B4QET3 A0A0A1XCX0 A1Z757 B4HR84 Q7JY97 K7J6K2 J3S999 A0A1W4VZF9 T1HKC6 A0A3B0J6V3 A0A1I8MLY4 A0A0K8RWA2 B3N9D8 A0A2A3EEH9 K1PPR1 A0A1I8MLY6 Q26373 B4J848 A0A0B7AUQ3 A0A232EFC2 A0A0K8V113 B4ME10 A0A1V4KMC1 W8C169 A0A088ARG8 A0A2B4RXJ6 B4P244 A0A0C9Q5W8 H9G6X4 A0A023F3G0 A0A194PQS4 A0A034V6B8 F6Q9E0 A0A0M4E6S9 A0A034V448 A0A1V4KMH6 A0A2I0LMR4 Q28464 A0A3M7QL57 B4MQY9 E9HAM9 G3VYP1 S4R979 A0A0P6JIK5 A0A151MZN9 A0A0N8CYE9
A0A182NZR1 A0A182H124 A0A182HPD9 Q17KA3 A0A084WCU3 A0A182TDL1 A0A182V452 A0A182X0X1 A0A182LB47 B0WAC3 A0A182WMB2 A0A1Q3F590 A0A182FY44 Q7Q9T5 A0A1B6H369 W5J446 A0A182NLW6 A0A182LSK5 A0A182RUX5 A0A182QNV4 A0A182YJ19 A0A1B6KN13 F6W9G4 A0A1B6EB20 A0A336L2S2 Q3C2L7 A0A336LWB1 A0A182JL00 A0A1J1J7Y8 A0A0L0BRI0 A0A1S3H867 A0A0A9WTY5 A0A2S2QTI6 V4A167 B3MG96 A0A0P8Y6L2 A0A0P4WEL0 A0A075F4M7 B4KQ29 A0A2S2NFH5 A0A2I0U149 A0A210PRW9 Q24443 Q28ZN6 J9K3D6 A0A0N1INB4 T1L328 B4GH10 B4QET3 A0A0A1XCX0 A1Z757 B4HR84 Q7JY97 K7J6K2 J3S999 A0A1W4VZF9 T1HKC6 A0A3B0J6V3 A0A1I8MLY4 A0A0K8RWA2 B3N9D8 A0A2A3EEH9 K1PPR1 A0A1I8MLY6 Q26373 B4J848 A0A0B7AUQ3 A0A232EFC2 A0A0K8V113 B4ME10 A0A1V4KMC1 W8C169 A0A088ARG8 A0A2B4RXJ6 B4P244 A0A0C9Q5W8 H9G6X4 A0A023F3G0 A0A194PQS4 A0A034V6B8 F6Q9E0 A0A0M4E6S9 A0A034V448 A0A1V4KMH6 A0A2I0LMR4 Q28464 A0A3M7QL57 B4MQY9 E9HAM9 G3VYP1 S4R979 A0A0P6JIK5 A0A151MZN9 A0A0N8CYE9
PDB
3UMV
E-value=3.17441e-118,
Score=1089
Ontologies
GO
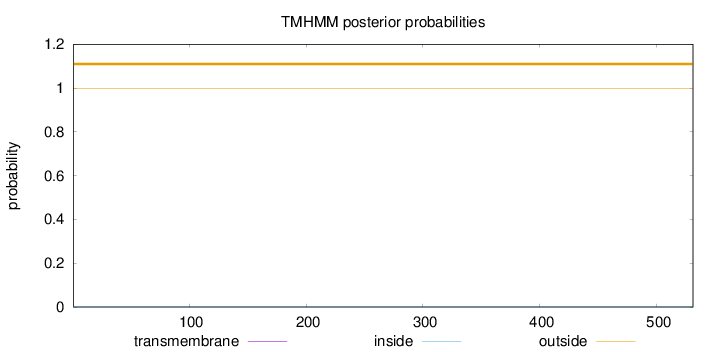
Topology
Length:
531
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00318
Exp number, first 60 AAs:
0.0019
Total prob of N-in:
0.00052
outside
1 - 531
Population Genetic Test Statistics
Pi
343.348244
Theta
198.284798
Tajima's D
2.055733
CLR
0.340918
CSRT
0.891205439728014
Interpretation
Uncertain
