Pre Gene Modal
BGIBMGA003584
Annotation
PREDICTED:_protein_tyrosine_phosphatase_type_IVA_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.062
Sequence
CDS
ATGAAGCAGAAGGACATTAGACCGGCGCCTTCCCTCATCGAGTACAAGGGGATGCGCTTCCTCATCACCGACCGCCCATCGGACGTGTCCATTCAGGCGTATCTGCAGGAGTTGAGGAAGCACAACGTGTGCACGGTGGTGCGCGTGTGCGAGCCGAGCTACGACACGGCGCCGCTGAAGGCCGAGAACATCGAGGTGCGCGACCTCGCCTACGACGACGGGACCTTCCCGCCAGCCAACGTCGTAGACGACTGGTTCGAGATCCTCAGAGATAAGGCGGAGAACAAGCCGGAGTCGGCGGTGGCGGTGCACTGCGTGGCGGGGCTGGGCCGCGCGCCCGTCATGGTCGCCATCGCGCTCATCGAGCTCGGCATGAAGTACGAAGAGGCCGTCGAGACCATCAGGGAGTAA
Protein
MKQKDIRPAPSLIEYKGMRFLITDRPSDVSIQAYLQELRKHNVCTVVRVCEPSYDTAPLKAENIEVRDLAYDDGTFPPANVVDDWFEILRDKAENKPESAVAVHCVAGLGRAPVMVAIALIELGMKYEEAVETIRE
Summary
Uniprot
A0A2H1W950
E3VWK8
A0A2A4JFN0
S4PYB1
A0A212EVF8
A0A1E1WAV0
+ More
A0A0N1PHW7 A0A194PP60 A0A0L7LP11 B4JPF3 B4M8Z1 B3MKX9 D3TNG4 A0A1A9VA60 A0A1A9W2J8 A0A1I8P6K0 Q29P39 A0A1I8N0S0 A0A0Q9X512 A0A0J9TPA2 B3N646 O61722 B4NXQ9 B4Q6Y6 Q95VY8 B4I202 A0A3B0K9J8 A0A1W4VJU7 W8AK94 A0A0K8TY85 A0A0K8TPC9 T1DIV3 B4MUF1 A0A1J1I1W8 A0A1L8DM13 A0A1Q3EUD9 A0A1Q3EUE9 Q17K22 A0A023EIY7 A0A182GQ11 A0A1Y1LQ27 A0A1L8EAC7 U5EWG9 V5GM23 A0A1Q3FYP3 A0A1Q3FYI3 A0A182VTF9 A0A182ILK6 A0A084VMH1 Q7KPK2 A0A023EH60 Q17K23 A0A0T6B0W2 A0A182NAL8 A0A3F2YQ53 A0A0C9PN91 T1E8F0 A0A2M4CTW0 A0A2M3ZFL3 A0A2M4A3C7 A0A182MBV4 A0A182R1B5 A0A182SLT9 A0A182VNA3 A0A182TH21 A0A182KWE9 A0A182X6A5 Q7PPM5 A0A182IBC7 A0A182YLQ8 A0A182PJH4 A0A1B0DLI0 R4WD48 N6TII5 A0A336LTY8 A0A1W4WQ38 A0A182M8F5 A0A1B6CMN8 A0A0A9X978 A0A232EUZ4 A0A182R3Z7 K7J561 A0A154PCD0 A0A0P5P3D5 A0A0P5MLX5 E9HWB4 A0A0P5DH36 A0A158NGW4 A0A0N8AUQ0 A0A195B6N5 E2ACE4 F4WZT6 A0A026VVP2 A0A151XD09 A0A151JBQ8 A0A1B6IA59 D6W854 A0A151IID1 A0A195FGP1 A0A1B6GB16
A0A0N1PHW7 A0A194PP60 A0A0L7LP11 B4JPF3 B4M8Z1 B3MKX9 D3TNG4 A0A1A9VA60 A0A1A9W2J8 A0A1I8P6K0 Q29P39 A0A1I8N0S0 A0A0Q9X512 A0A0J9TPA2 B3N646 O61722 B4NXQ9 B4Q6Y6 Q95VY8 B4I202 A0A3B0K9J8 A0A1W4VJU7 W8AK94 A0A0K8TY85 A0A0K8TPC9 T1DIV3 B4MUF1 A0A1J1I1W8 A0A1L8DM13 A0A1Q3EUD9 A0A1Q3EUE9 Q17K22 A0A023EIY7 A0A182GQ11 A0A1Y1LQ27 A0A1L8EAC7 U5EWG9 V5GM23 A0A1Q3FYP3 A0A1Q3FYI3 A0A182VTF9 A0A182ILK6 A0A084VMH1 Q7KPK2 A0A023EH60 Q17K23 A0A0T6B0W2 A0A182NAL8 A0A3F2YQ53 A0A0C9PN91 T1E8F0 A0A2M4CTW0 A0A2M3ZFL3 A0A2M4A3C7 A0A182MBV4 A0A182R1B5 A0A182SLT9 A0A182VNA3 A0A182TH21 A0A182KWE9 A0A182X6A5 Q7PPM5 A0A182IBC7 A0A182YLQ8 A0A182PJH4 A0A1B0DLI0 R4WD48 N6TII5 A0A336LTY8 A0A1W4WQ38 A0A182M8F5 A0A1B6CMN8 A0A0A9X978 A0A232EUZ4 A0A182R3Z7 K7J561 A0A154PCD0 A0A0P5P3D5 A0A0P5MLX5 E9HWB4 A0A0P5DH36 A0A158NGW4 A0A0N8AUQ0 A0A195B6N5 E2ACE4 F4WZT6 A0A026VVP2 A0A151XD09 A0A151JBQ8 A0A1B6IA59 D6W854 A0A151IID1 A0A195FGP1 A0A1B6GB16
Pubmed
23622113
22118469
26354079
26227816
17994087
18057021
+ More
20353571 15632085 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 26369729 24330624 17510324 24945155 26483478 28004739 24438588 20966253 12364791 14747013 17210077 25244985 23691247 23537049 25401762 26823975 28648823 20075255 21292972 21347285 20798317 21719571 24508170 30249741 18362917 19820115
20353571 15632085 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 26369729 24330624 17510324 24945155 26483478 28004739 24438588 20966253 12364791 14747013 17210077 25244985 23691247 23537049 25401762 26823975 28648823 20075255 21292972 21347285 20798317 21719571 24508170 30249741 18362917 19820115
EMBL
ODYU01007111
SOQ49598.1
HQ292704
ADP20217.1
NWSH01001547
PCG70895.1
+ More
GAIX01003598 JAA88962.1 AGBW02012194 OWR45476.1 GDQN01007898 GDQN01006932 GDQN01000876 JAT83156.1 JAT84122.1 JAT90178.1 KQ461187 KPJ07336.1 KQ459597 KPI95092.1 JTDY01000442 KOB77150.1 CH916372 EDV98783.1 CH940654 EDW57667.1 KRF77874.1 KRF77876.1 CH902620 EDV30637.1 EZ422966 ADD19242.1 CH379058 EAL34454.1 CH933807 KRG03215.1 CM002910 KMY90475.1 KMY90478.1 CH954177 EDV58084.1 KQS70240.1 KQS70241.1 AE014134 AY071505 AAF53506.2 AAL49127.1 AAN10934.1 AGB93022.1 AGB93023.1 CM000157 EDW89684.1 KRJ98652.1 KRJ98653.1 KRJ98654.1 CM000361 EDX05194.1 AF390535 AAL26988.1 CH480820 EDW54559.1 OUUW01000020 SPP89372.1 GAMC01021422 JAB85133.1 GDHF01032865 GDHF01016893 JAI19449.1 JAI35421.1 GDAI01001595 JAI16008.1 GALA01000731 JAA94121.1 CH963857 EDW76077.1 CVRI01000038 CRK94285.1 GFDF01006669 JAV07415.1 GFDL01016111 JAV18934.1 GFDL01016112 JAV18933.1 CH477228 EAT47069.1 GAPW01004979 GAPW01004964 JAC08634.1 JXUM01079760 JXUM01079761 KQ563144 KXJ74397.1 GEZM01049871 GEZM01049870 GEZM01049869 JAV75794.1 GFDG01003079 JAV15720.1 GANO01001420 JAB58451.1 GALX01003337 JAB65129.1 GFDL01002397 JAV32648.1 GFDL01002400 JAV32645.1 ATLV01014596 KE524975 KFB39165.1 AF063902 AAC16552.1 GAPW01004895 JAC08703.1 EAT47070.1 LJIG01016304 KRT81004.1 GBYB01002628 JAG72395.1 GAMD01002237 JAA99353.1 GGFL01004433 MBW68611.1 GGFM01006548 MBW27299.1 GGFK01001934 MBW35255.1 AXCM01006822 AXCN02002186 AAAB01008944 EAA10162.4 APCN01000667 AJVK01006666 AJVK01006667 AK417388 BAN20603.1 APGK01024770 KB740562 KB632399 ENN80244.1 ERL94850.1 UFQT01000121 SSX20441.1 AXCM01016374 GEDC01030758 GEDC01030707 GEDC01023026 GEDC01022606 GEDC01020343 GEDC01014728 GEDC01014330 GEDC01014143 JAS06540.1 JAS06591.1 JAS14272.1 JAS14692.1 JAS16955.1 JAS22570.1 JAS22968.1 JAS23155.1 GBHO01036403 GBHO01036400 GBHO01027065 GBHO01027056 GBRD01000951 GBRD01000950 GDHC01020024 GDHC01002290 JAG07201.1 JAG07204.1 JAG16539.1 JAG16548.1 JAG64870.1 JAP98604.1 JAQ16339.1 NNAY01002071 OXU22166.1 AAZX01014434 KQ434870 KZC09502.1 GDIQ01140647 JAL11079.1 GDIQ01157490 JAK94235.1 GL732911 EFX63968.1 GDIP01156333 GDIQ01095949 JAJ67069.1 JAL55777.1 ADTU01015286 GDIQ01241059 JAK10666.1 KQ976574 KYM80178.1 GL438491 EFN68934.1 GL888480 EGI60361.1 KK107770 QOIP01000012 EZA47827.1 RLU15855.1 KQ982294 KYQ58264.1 KQ979114 KYN22591.1 GECU01023933 GECU01010776 JAS83773.1 JAS96930.1 KQ971307 EFA11186.1 KQ977492 KYN02303.1 KQ981625 KYN39169.1 GECZ01025399 GECZ01010148 GECZ01006113 JAS44370.1 JAS59621.1 JAS63656.1
GAIX01003598 JAA88962.1 AGBW02012194 OWR45476.1 GDQN01007898 GDQN01006932 GDQN01000876 JAT83156.1 JAT84122.1 JAT90178.1 KQ461187 KPJ07336.1 KQ459597 KPI95092.1 JTDY01000442 KOB77150.1 CH916372 EDV98783.1 CH940654 EDW57667.1 KRF77874.1 KRF77876.1 CH902620 EDV30637.1 EZ422966 ADD19242.1 CH379058 EAL34454.1 CH933807 KRG03215.1 CM002910 KMY90475.1 KMY90478.1 CH954177 EDV58084.1 KQS70240.1 KQS70241.1 AE014134 AY071505 AAF53506.2 AAL49127.1 AAN10934.1 AGB93022.1 AGB93023.1 CM000157 EDW89684.1 KRJ98652.1 KRJ98653.1 KRJ98654.1 CM000361 EDX05194.1 AF390535 AAL26988.1 CH480820 EDW54559.1 OUUW01000020 SPP89372.1 GAMC01021422 JAB85133.1 GDHF01032865 GDHF01016893 JAI19449.1 JAI35421.1 GDAI01001595 JAI16008.1 GALA01000731 JAA94121.1 CH963857 EDW76077.1 CVRI01000038 CRK94285.1 GFDF01006669 JAV07415.1 GFDL01016111 JAV18934.1 GFDL01016112 JAV18933.1 CH477228 EAT47069.1 GAPW01004979 GAPW01004964 JAC08634.1 JXUM01079760 JXUM01079761 KQ563144 KXJ74397.1 GEZM01049871 GEZM01049870 GEZM01049869 JAV75794.1 GFDG01003079 JAV15720.1 GANO01001420 JAB58451.1 GALX01003337 JAB65129.1 GFDL01002397 JAV32648.1 GFDL01002400 JAV32645.1 ATLV01014596 KE524975 KFB39165.1 AF063902 AAC16552.1 GAPW01004895 JAC08703.1 EAT47070.1 LJIG01016304 KRT81004.1 GBYB01002628 JAG72395.1 GAMD01002237 JAA99353.1 GGFL01004433 MBW68611.1 GGFM01006548 MBW27299.1 GGFK01001934 MBW35255.1 AXCM01006822 AXCN02002186 AAAB01008944 EAA10162.4 APCN01000667 AJVK01006666 AJVK01006667 AK417388 BAN20603.1 APGK01024770 KB740562 KB632399 ENN80244.1 ERL94850.1 UFQT01000121 SSX20441.1 AXCM01016374 GEDC01030758 GEDC01030707 GEDC01023026 GEDC01022606 GEDC01020343 GEDC01014728 GEDC01014330 GEDC01014143 JAS06540.1 JAS06591.1 JAS14272.1 JAS14692.1 JAS16955.1 JAS22570.1 JAS22968.1 JAS23155.1 GBHO01036403 GBHO01036400 GBHO01027065 GBHO01027056 GBRD01000951 GBRD01000950 GDHC01020024 GDHC01002290 JAG07201.1 JAG07204.1 JAG16539.1 JAG16548.1 JAG64870.1 JAP98604.1 JAQ16339.1 NNAY01002071 OXU22166.1 AAZX01014434 KQ434870 KZC09502.1 GDIQ01140647 JAL11079.1 GDIQ01157490 JAK94235.1 GL732911 EFX63968.1 GDIP01156333 GDIQ01095949 JAJ67069.1 JAL55777.1 ADTU01015286 GDIQ01241059 JAK10666.1 KQ976574 KYM80178.1 GL438491 EFN68934.1 GL888480 EGI60361.1 KK107770 QOIP01000012 EZA47827.1 RLU15855.1 KQ982294 KYQ58264.1 KQ979114 KYN22591.1 GECU01023933 GECU01010776 JAS83773.1 JAS96930.1 KQ971307 EFA11186.1 KQ977492 KYN02303.1 KQ981625 KYN39169.1 GECZ01025399 GECZ01010148 GECZ01006113 JAS44370.1 JAS59621.1 JAS63656.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
UP000001070
+ More
UP000008792 UP000007801 UP000078200 UP000091820 UP000095300 UP000001819 UP000095301 UP000009192 UP000008711 UP000000803 UP000002282 UP000000304 UP000001292 UP000268350 UP000192221 UP000007798 UP000183832 UP000008820 UP000069940 UP000249989 UP000075920 UP000075880 UP000030765 UP000075884 UP000069272 UP000075883 UP000075886 UP000075901 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000076408 UP000075885 UP000092462 UP000019118 UP000030742 UP000192223 UP000215335 UP000075900 UP000002358 UP000076502 UP000000305 UP000005205 UP000078540 UP000000311 UP000007755 UP000053097 UP000279307 UP000075809 UP000078492 UP000007266 UP000078542 UP000078541
UP000008792 UP000007801 UP000078200 UP000091820 UP000095300 UP000001819 UP000095301 UP000009192 UP000008711 UP000000803 UP000002282 UP000000304 UP000001292 UP000268350 UP000192221 UP000007798 UP000183832 UP000008820 UP000069940 UP000249989 UP000075920 UP000075880 UP000030765 UP000075884 UP000069272 UP000075883 UP000075886 UP000075901 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000076408 UP000075885 UP000092462 UP000019118 UP000030742 UP000192223 UP000215335 UP000075900 UP000002358 UP000076502 UP000000305 UP000005205 UP000078540 UP000000311 UP000007755 UP000053097 UP000279307 UP000075809 UP000078492 UP000007266 UP000078542 UP000078541
Pfam
PF00782 DSPc
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
A0A2H1W950
E3VWK8
A0A2A4JFN0
S4PYB1
A0A212EVF8
A0A1E1WAV0
+ More
A0A0N1PHW7 A0A194PP60 A0A0L7LP11 B4JPF3 B4M8Z1 B3MKX9 D3TNG4 A0A1A9VA60 A0A1A9W2J8 A0A1I8P6K0 Q29P39 A0A1I8N0S0 A0A0Q9X512 A0A0J9TPA2 B3N646 O61722 B4NXQ9 B4Q6Y6 Q95VY8 B4I202 A0A3B0K9J8 A0A1W4VJU7 W8AK94 A0A0K8TY85 A0A0K8TPC9 T1DIV3 B4MUF1 A0A1J1I1W8 A0A1L8DM13 A0A1Q3EUD9 A0A1Q3EUE9 Q17K22 A0A023EIY7 A0A182GQ11 A0A1Y1LQ27 A0A1L8EAC7 U5EWG9 V5GM23 A0A1Q3FYP3 A0A1Q3FYI3 A0A182VTF9 A0A182ILK6 A0A084VMH1 Q7KPK2 A0A023EH60 Q17K23 A0A0T6B0W2 A0A182NAL8 A0A3F2YQ53 A0A0C9PN91 T1E8F0 A0A2M4CTW0 A0A2M3ZFL3 A0A2M4A3C7 A0A182MBV4 A0A182R1B5 A0A182SLT9 A0A182VNA3 A0A182TH21 A0A182KWE9 A0A182X6A5 Q7PPM5 A0A182IBC7 A0A182YLQ8 A0A182PJH4 A0A1B0DLI0 R4WD48 N6TII5 A0A336LTY8 A0A1W4WQ38 A0A182M8F5 A0A1B6CMN8 A0A0A9X978 A0A232EUZ4 A0A182R3Z7 K7J561 A0A154PCD0 A0A0P5P3D5 A0A0P5MLX5 E9HWB4 A0A0P5DH36 A0A158NGW4 A0A0N8AUQ0 A0A195B6N5 E2ACE4 F4WZT6 A0A026VVP2 A0A151XD09 A0A151JBQ8 A0A1B6IA59 D6W854 A0A151IID1 A0A195FGP1 A0A1B6GB16
A0A0N1PHW7 A0A194PP60 A0A0L7LP11 B4JPF3 B4M8Z1 B3MKX9 D3TNG4 A0A1A9VA60 A0A1A9W2J8 A0A1I8P6K0 Q29P39 A0A1I8N0S0 A0A0Q9X512 A0A0J9TPA2 B3N646 O61722 B4NXQ9 B4Q6Y6 Q95VY8 B4I202 A0A3B0K9J8 A0A1W4VJU7 W8AK94 A0A0K8TY85 A0A0K8TPC9 T1DIV3 B4MUF1 A0A1J1I1W8 A0A1L8DM13 A0A1Q3EUD9 A0A1Q3EUE9 Q17K22 A0A023EIY7 A0A182GQ11 A0A1Y1LQ27 A0A1L8EAC7 U5EWG9 V5GM23 A0A1Q3FYP3 A0A1Q3FYI3 A0A182VTF9 A0A182ILK6 A0A084VMH1 Q7KPK2 A0A023EH60 Q17K23 A0A0T6B0W2 A0A182NAL8 A0A3F2YQ53 A0A0C9PN91 T1E8F0 A0A2M4CTW0 A0A2M3ZFL3 A0A2M4A3C7 A0A182MBV4 A0A182R1B5 A0A182SLT9 A0A182VNA3 A0A182TH21 A0A182KWE9 A0A182X6A5 Q7PPM5 A0A182IBC7 A0A182YLQ8 A0A182PJH4 A0A1B0DLI0 R4WD48 N6TII5 A0A336LTY8 A0A1W4WQ38 A0A182M8F5 A0A1B6CMN8 A0A0A9X978 A0A232EUZ4 A0A182R3Z7 K7J561 A0A154PCD0 A0A0P5P3D5 A0A0P5MLX5 E9HWB4 A0A0P5DH36 A0A158NGW4 A0A0N8AUQ0 A0A195B6N5 E2ACE4 F4WZT6 A0A026VVP2 A0A151XD09 A0A151JBQ8 A0A1B6IA59 D6W854 A0A151IID1 A0A195FGP1 A0A1B6GB16
PDB
5K22
E-value=3.18523e-39,
Score=399
Ontologies
GO
Topology
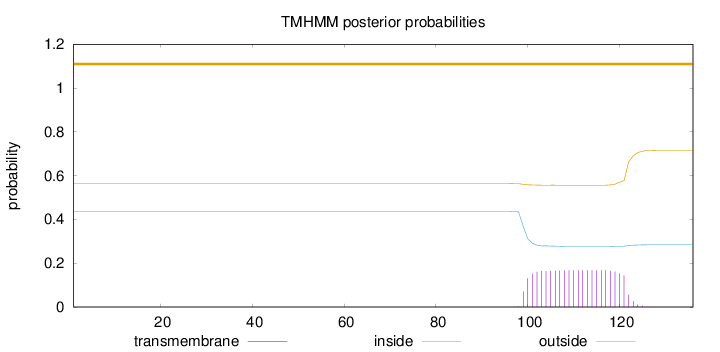
Length:
136
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.74137
Exp number, first 60 AAs:
0
Total prob of N-in:
0.43653
outside
1 - 136
Population Genetic Test Statistics
Pi
147.264875
Theta
146.360256
Tajima's D
0.001058
CLR
1.045441
CSRT
0.365131743412829
Interpretation
Uncertain
