Gene
KWMTBOMO02992 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003583
Annotation
PREDICTED:_proteasome_inhibitor_PI31_subunit_[Plutella_xylostella]
Full name
Proteasome inhibitor PI31 subunit
Location in the cell
Cytoplasmic Reliability : 1.621 Nuclear Reliability : 1.252
Sequence
CDS
ATGGCATCAGACCCGCTGTTTGGCTGGGATTTAACATTCAAAACAATTGAGAGGGATATAAAAAAACAGTCTGACATAATAATTGCTTTTATTCACTGGAATTTAACAAGGAGGGGTTTTCGAAGCATCGGCTTAAGTGATGAGAGAACAATCACTGGCGATGAACCTAAAAGTGAGCTACTTCCTGAAGGATGGAACGACACTGAGAACTACAGAATAAGATATGTTCTCGAATCTAAACTCTATATTCTACATGGCTTAAACACTGATGGAAATTTAATTGTAAATTTAATGAGATCTGAAGACCTTGCTGTTACGAACATAGGTGTAAAGATAGACGAAATACTGAAAGAATCAAATGGAACCATAGACGTCATGATGCCGAATTACAAAGATTTCATTTTTGTTATAAAGCGAGATCTAATTGATAGCATCACAGACAAGCCAACAGCTACTTCGGAAACACAGACTGCATCAGATCATTCAAACACTAGAAGGCCTTCAGATGATCCACTGAGAATTCCTCCCAGACCCAGACCCGGAGTTAATCCCGATTCACAAGATCTATGGGCAGTTCCTCCTGGCCGTAACGTTGGCCATTCAGATCTCGATCCGTTCTCTCCTTTCGGGGGTGGAATGATATTTAACCCGTTTGCTCCACGTCGAGACATTGAGAACCCAGGCCTGGGTGTTCCAGGTGGATTGCCAAGAGCTGCGGTTCCCCCGGGGGCACGTTTCGATCCGTTTGCGCCCCCCGGTGTTGGTGAACCCATCCCGGGAAGAAGGCCACCTCCTGACGCTGATCACCTTCCACCTCCAGGAGGCTTTAATGAGAATATGTTCATGTAA
Protein
MASDPLFGWDLTFKTIERDIKKQSDIIIAFIHWNLTRRGFRSIGLSDERTITGDEPKSELLPEGWNDTENYRIRYVLESKLYILHGLNTDGNLIVNLMRSEDLAVTNIGVKIDEILKESNGTIDVMMPNYKDFIFVIKRDLIDSITDKPTATSETQTASDHSNTRRPSDDPLRIPPRPRPGVNPDSQDLWAVPPGRNVGHSDLDPFSPFGGGMIFNPFAPRRDIENPGLGVPGGLPRAAVPPGARFDPFAPPGVGEPIPGRRPPPDADHLPPPGGFNENMFM
Summary
Description
Plays an important role in the control of proteasome function. Inhibits the hydrolysis of protein and peptide substrates by the 20S proteasome. Enhances 26S proteasome function by promoting its assembly through the interaction with the assembly chaperones PSMD9 and PMSD5. Functions together with ntc to control non-apoptotic caspase activation during sperm individualization. In testis, is required for proper protein degradation and germline cell cycle progression.
Subunit
Interacts with ntc. Interacts with Tnks (via ANK repeats). Interacts with 20S proteasome subunits Prosalpha1, Prosalpha3, Prosalpha4 and Prosalpha6, but not with Prosalpha2 and Prosalpha5. The interaction with Prosalpha4 is reduced by PI31 ADP-ribosylation. Interacts with the 19S assembly chaperones PSMD5 and PSMD9; these interactions are increased by PI31 ADP-ribosylation.
Similarity
Belongs to the proteasome inhibitor PI31 family.
Keywords
ADP-ribosylation
Complete proteome
Cytoplasm
Developmental protein
Phosphoprotein
Proteasome
Reference proteome
Feature
chain Proteasome inhibitor PI31 subunit
Uniprot
H9J246
A0A2A4JHI1
A0A2H1WYP4
S4NVB2
A0A194PVD6
A0A0N1PFH8
+ More
A0A1E1WTV4 A0A0L7LPP5 A0A067RET4 E0W495 A0A1B6H0N6 A0A2J7Q6J5 A0A1B6LA54 A0A0A9YXU3 A0A154PCF6 A0A0V0G6M4 R4WS17 A0A232EUZ2 A0A2P8YHL3 K7J560 A0A224XHR2 A0A069DYC4 E2ACE3 A0A151JBV8 F4WZT7 A0A0M8ZNL0 A0A151IIQ7 A0A151XD28 A0A195B7F3 A0A158NGW5 E9J8A2 A0A310SCG5 A0A026VWC7 A0A1Q3FGZ4 B0VZG5 A0A1L8DSK9 A0A2A3E7I2 Q17LA8 A0A023EM48 A0A088AVV4 A0A195FER9 A0A0P4VIE1 R4G7Y8 U5EX73 V5I9I2 A0A0L7R3Q6 A0A1B6D1P0 D2A288 E2BWD8 A0A1W4WAU1 A0A0C9QED1 A0A1S3DGC0 A0A1B0EU12 N6SVV0 A2I453 A0A1Y1MTJ4 A0A182XYT0 A0A182R7E5 A0A084WN61 A0A0T6B2N9 A0A1L8EI46 A0A336K604 A0A182JA66 A0A182JT18 A0A1A9UXY4 A0A0L0CDF6 B5DGA4 A0A1I8P461 A0A0R4II16 A0A1A9W0Y6 T1GIL4 A0A2M4BXA7 A0A182WJB2 K1QP56 A0A0K8TRZ7 A0A1A9UXY7 A0A3N0YXF6 B4QC54 A0A1W4Z1S7 C0PUJ2 B4J9W0 A0A1A9UU06 B3MG42 A0A0B8RU44 A0A182MH61 A0A1A9ZUX4 A0A0B4KEK7 Q9V637 A0A060WKI2 A0A2M4CQP4 A0A1J1IIW7 A0A182FHI1 A0A1A9WE05 A0A0K8U478
A0A1E1WTV4 A0A0L7LPP5 A0A067RET4 E0W495 A0A1B6H0N6 A0A2J7Q6J5 A0A1B6LA54 A0A0A9YXU3 A0A154PCF6 A0A0V0G6M4 R4WS17 A0A232EUZ2 A0A2P8YHL3 K7J560 A0A224XHR2 A0A069DYC4 E2ACE3 A0A151JBV8 F4WZT7 A0A0M8ZNL0 A0A151IIQ7 A0A151XD28 A0A195B7F3 A0A158NGW5 E9J8A2 A0A310SCG5 A0A026VWC7 A0A1Q3FGZ4 B0VZG5 A0A1L8DSK9 A0A2A3E7I2 Q17LA8 A0A023EM48 A0A088AVV4 A0A195FER9 A0A0P4VIE1 R4G7Y8 U5EX73 V5I9I2 A0A0L7R3Q6 A0A1B6D1P0 D2A288 E2BWD8 A0A1W4WAU1 A0A0C9QED1 A0A1S3DGC0 A0A1B0EU12 N6SVV0 A2I453 A0A1Y1MTJ4 A0A182XYT0 A0A182R7E5 A0A084WN61 A0A0T6B2N9 A0A1L8EI46 A0A336K604 A0A182JA66 A0A182JT18 A0A1A9UXY4 A0A0L0CDF6 B5DGA4 A0A1I8P461 A0A0R4II16 A0A1A9W0Y6 T1GIL4 A0A2M4BXA7 A0A182WJB2 K1QP56 A0A0K8TRZ7 A0A1A9UXY7 A0A3N0YXF6 B4QC54 A0A1W4Z1S7 C0PUJ2 B4J9W0 A0A1A9UU06 B3MG42 A0A0B8RU44 A0A182MH61 A0A1A9ZUX4 A0A0B4KEK7 Q9V637 A0A060WKI2 A0A2M4CQP4 A0A1J1IIW7 A0A182FHI1 A0A1A9WE05 A0A0K8U478
Pubmed
19121390
23622113
26354079
26227816
24845553
20566863
+ More
25401762 26823975 23691247 28648823 29403074 20075255 26334808 20798317 21719571 21347285 21282665 24508170 30249741 17510324 24945155 27129103 18362917 19820115 23537049 28004739 25244985 24438588 26108605 19878547 20433749 23594743 22992520 26369729 17994087 22936249 25476704 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 17372656 18327897 21529711 23622245 24755649
25401762 26823975 23691247 28648823 29403074 20075255 26334808 20798317 21719571 21347285 21282665 24508170 30249741 17510324 24945155 27129103 18362917 19820115 23537049 28004739 25244985 24438588 26108605 19878547 20433749 23594743 22992520 26369729 17994087 22936249 25476704 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 17372656 18327897 21529711 23622245 24755649
EMBL
BABH01007437
BABH01007438
NWSH01001547
PCG70893.1
ODYU01012083
SOQ58195.1
+ More
GAIX01013017 JAA79543.1 KQ459597 KPI95090.1 KQ461187 KPJ07334.1 GDQN01000646 JAT90408.1 JTDY01000442 KOB77151.1 KK852549 KDR21548.1 DS235886 EEB20451.1 GECZ01001523 JAS68246.1 NEVH01017466 PNF24211.1 GEBQ01019404 JAT20573.1 GBHO01007688 GBHO01007686 GDHC01004441 JAG35916.1 JAG35918.1 JAQ14188.1 KQ434870 KZC09503.1 GECL01002462 JAP03662.1 AK417492 BAN20707.1 NNAY01002071 OXU22164.1 PYGN01000589 PSN43741.1 AAZX01014434 GFTR01004401 JAW12025.1 GBGD01002490 JAC86399.1 GL438491 EFN68933.1 KQ979114 KYN22592.1 GL888480 EGI60362.1 KQ435969 KOX67905.1 KQ977492 KYN02302.1 KQ982294 KYQ58265.1 KQ976574 KYM80177.1 ADTU01015286 GL768914 EFZ10952.1 KQ763551 OAD55004.1 KK107770 QOIP01000012 EZA47826.1 RLU15854.1 GFDL01008228 JAV26817.1 DS231814 EDS31702.1 GFDF01004655 JAV09429.1 KZ288342 PBC27723.1 CH477217 EAT47467.1 GAPW01003237 JAC10361.1 KQ981625 KYN39170.1 GDKW01002047 JAI54548.1 ACPB03021088 GAHY01001897 JAA75613.1 GANO01001075 JAB58796.1 GALX01002890 JAB65576.1 KQ414663 KOC65517.1 GEDC01017676 JAS19622.1 KQ971338 EFA02057.1 GL451103 EFN80042.1 GBYB01003015 GBYB01003017 GBYB01012768 GBYB01012769 JAG72782.1 JAG72784.1 JAG82535.1 JAG82536.1 AJWK01011174 APGK01054419 KB741250 KB632390 ENN71889.1 ERL94502.1 EF070556 ABM55622.1 GEZM01021637 JAV88889.1 ATLV01024569 KE525352 KFB51655.1 LJIG01016110 KRT81586.1 GFDG01000474 JAV18325.1 UFQS01000040 UFQT01000040 SSW98163.1 SSX18549.1 JRES01000655 KNC29504.1 BT043663 ACH70778.1 BX001037 CAQQ02001161 GGFJ01008522 MBW57663.1 JH816890 EKC23326.1 GDAI01000481 JAI17122.1 RJVU01019434 ROL50623.1 CM000362 CM002911 EDX06685.1 KMY93062.1 BT072465 ACN60283.1 CH916367 EDW01524.1 CH902619 EDV36737.1 GBSH01001980 JAG67046.1 AXCM01001831 AE013599 AGB93438.1 AGB93440.1 AY058250 FR904595 CDQ67763.1 GGFL01003407 MBW67585.1 CVRI01000051 CRK99484.1 GDHF01031008 JAI21306.1
GAIX01013017 JAA79543.1 KQ459597 KPI95090.1 KQ461187 KPJ07334.1 GDQN01000646 JAT90408.1 JTDY01000442 KOB77151.1 KK852549 KDR21548.1 DS235886 EEB20451.1 GECZ01001523 JAS68246.1 NEVH01017466 PNF24211.1 GEBQ01019404 JAT20573.1 GBHO01007688 GBHO01007686 GDHC01004441 JAG35916.1 JAG35918.1 JAQ14188.1 KQ434870 KZC09503.1 GECL01002462 JAP03662.1 AK417492 BAN20707.1 NNAY01002071 OXU22164.1 PYGN01000589 PSN43741.1 AAZX01014434 GFTR01004401 JAW12025.1 GBGD01002490 JAC86399.1 GL438491 EFN68933.1 KQ979114 KYN22592.1 GL888480 EGI60362.1 KQ435969 KOX67905.1 KQ977492 KYN02302.1 KQ982294 KYQ58265.1 KQ976574 KYM80177.1 ADTU01015286 GL768914 EFZ10952.1 KQ763551 OAD55004.1 KK107770 QOIP01000012 EZA47826.1 RLU15854.1 GFDL01008228 JAV26817.1 DS231814 EDS31702.1 GFDF01004655 JAV09429.1 KZ288342 PBC27723.1 CH477217 EAT47467.1 GAPW01003237 JAC10361.1 KQ981625 KYN39170.1 GDKW01002047 JAI54548.1 ACPB03021088 GAHY01001897 JAA75613.1 GANO01001075 JAB58796.1 GALX01002890 JAB65576.1 KQ414663 KOC65517.1 GEDC01017676 JAS19622.1 KQ971338 EFA02057.1 GL451103 EFN80042.1 GBYB01003015 GBYB01003017 GBYB01012768 GBYB01012769 JAG72782.1 JAG72784.1 JAG82535.1 JAG82536.1 AJWK01011174 APGK01054419 KB741250 KB632390 ENN71889.1 ERL94502.1 EF070556 ABM55622.1 GEZM01021637 JAV88889.1 ATLV01024569 KE525352 KFB51655.1 LJIG01016110 KRT81586.1 GFDG01000474 JAV18325.1 UFQS01000040 UFQT01000040 SSW98163.1 SSX18549.1 JRES01000655 KNC29504.1 BT043663 ACH70778.1 BX001037 CAQQ02001161 GGFJ01008522 MBW57663.1 JH816890 EKC23326.1 GDAI01000481 JAI17122.1 RJVU01019434 ROL50623.1 CM000362 CM002911 EDX06685.1 KMY93062.1 BT072465 ACN60283.1 CH916367 EDW01524.1 CH902619 EDV36737.1 GBSH01001980 JAG67046.1 AXCM01001831 AE013599 AGB93438.1 AGB93440.1 AY058250 FR904595 CDQ67763.1 GGFL01003407 MBW67585.1 CVRI01000051 CRK99484.1 GDHF01031008 JAI21306.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000027135
+ More
UP000009046 UP000235965 UP000076502 UP000215335 UP000245037 UP000002358 UP000000311 UP000078492 UP000007755 UP000053105 UP000078542 UP000075809 UP000078540 UP000005205 UP000053097 UP000279307 UP000002320 UP000242457 UP000008820 UP000005203 UP000078541 UP000015103 UP000053825 UP000007266 UP000008237 UP000192223 UP000079169 UP000092461 UP000019118 UP000030742 UP000076408 UP000075900 UP000030765 UP000075880 UP000075881 UP000078200 UP000037069 UP000087266 UP000095300 UP000000437 UP000091820 UP000015102 UP000075920 UP000005408 UP000000304 UP000192224 UP000001070 UP000007801 UP000075883 UP000092445 UP000000803 UP000193380 UP000183832 UP000069272
UP000009046 UP000235965 UP000076502 UP000215335 UP000245037 UP000002358 UP000000311 UP000078492 UP000007755 UP000053105 UP000078542 UP000075809 UP000078540 UP000005205 UP000053097 UP000279307 UP000002320 UP000242457 UP000008820 UP000005203 UP000078541 UP000015103 UP000053825 UP000007266 UP000008237 UP000192223 UP000079169 UP000092461 UP000019118 UP000030742 UP000076408 UP000075900 UP000030765 UP000075880 UP000075881 UP000078200 UP000037069 UP000087266 UP000095300 UP000000437 UP000091820 UP000015102 UP000075920 UP000005408 UP000000304 UP000192224 UP000001070 UP000007801 UP000075883 UP000092445 UP000000803 UP000193380 UP000183832 UP000069272
ProteinModelPortal
H9J246
A0A2A4JHI1
A0A2H1WYP4
S4NVB2
A0A194PVD6
A0A0N1PFH8
+ More
A0A1E1WTV4 A0A0L7LPP5 A0A067RET4 E0W495 A0A1B6H0N6 A0A2J7Q6J5 A0A1B6LA54 A0A0A9YXU3 A0A154PCF6 A0A0V0G6M4 R4WS17 A0A232EUZ2 A0A2P8YHL3 K7J560 A0A224XHR2 A0A069DYC4 E2ACE3 A0A151JBV8 F4WZT7 A0A0M8ZNL0 A0A151IIQ7 A0A151XD28 A0A195B7F3 A0A158NGW5 E9J8A2 A0A310SCG5 A0A026VWC7 A0A1Q3FGZ4 B0VZG5 A0A1L8DSK9 A0A2A3E7I2 Q17LA8 A0A023EM48 A0A088AVV4 A0A195FER9 A0A0P4VIE1 R4G7Y8 U5EX73 V5I9I2 A0A0L7R3Q6 A0A1B6D1P0 D2A288 E2BWD8 A0A1W4WAU1 A0A0C9QED1 A0A1S3DGC0 A0A1B0EU12 N6SVV0 A2I453 A0A1Y1MTJ4 A0A182XYT0 A0A182R7E5 A0A084WN61 A0A0T6B2N9 A0A1L8EI46 A0A336K604 A0A182JA66 A0A182JT18 A0A1A9UXY4 A0A0L0CDF6 B5DGA4 A0A1I8P461 A0A0R4II16 A0A1A9W0Y6 T1GIL4 A0A2M4BXA7 A0A182WJB2 K1QP56 A0A0K8TRZ7 A0A1A9UXY7 A0A3N0YXF6 B4QC54 A0A1W4Z1S7 C0PUJ2 B4J9W0 A0A1A9UU06 B3MG42 A0A0B8RU44 A0A182MH61 A0A1A9ZUX4 A0A0B4KEK7 Q9V637 A0A060WKI2 A0A2M4CQP4 A0A1J1IIW7 A0A182FHI1 A0A1A9WE05 A0A0K8U478
A0A1E1WTV4 A0A0L7LPP5 A0A067RET4 E0W495 A0A1B6H0N6 A0A2J7Q6J5 A0A1B6LA54 A0A0A9YXU3 A0A154PCF6 A0A0V0G6M4 R4WS17 A0A232EUZ2 A0A2P8YHL3 K7J560 A0A224XHR2 A0A069DYC4 E2ACE3 A0A151JBV8 F4WZT7 A0A0M8ZNL0 A0A151IIQ7 A0A151XD28 A0A195B7F3 A0A158NGW5 E9J8A2 A0A310SCG5 A0A026VWC7 A0A1Q3FGZ4 B0VZG5 A0A1L8DSK9 A0A2A3E7I2 Q17LA8 A0A023EM48 A0A088AVV4 A0A195FER9 A0A0P4VIE1 R4G7Y8 U5EX73 V5I9I2 A0A0L7R3Q6 A0A1B6D1P0 D2A288 E2BWD8 A0A1W4WAU1 A0A0C9QED1 A0A1S3DGC0 A0A1B0EU12 N6SVV0 A2I453 A0A1Y1MTJ4 A0A182XYT0 A0A182R7E5 A0A084WN61 A0A0T6B2N9 A0A1L8EI46 A0A336K604 A0A182JA66 A0A182JT18 A0A1A9UXY4 A0A0L0CDF6 B5DGA4 A0A1I8P461 A0A0R4II16 A0A1A9W0Y6 T1GIL4 A0A2M4BXA7 A0A182WJB2 K1QP56 A0A0K8TRZ7 A0A1A9UXY7 A0A3N0YXF6 B4QC54 A0A1W4Z1S7 C0PUJ2 B4J9W0 A0A1A9UU06 B3MG42 A0A0B8RU44 A0A182MH61 A0A1A9ZUX4 A0A0B4KEK7 Q9V637 A0A060WKI2 A0A2M4CQP4 A0A1J1IIW7 A0A182FHI1 A0A1A9WE05 A0A0K8U478
PDB
4OUH
E-value=2.90965e-08,
Score=137
Ontologies
KEGG
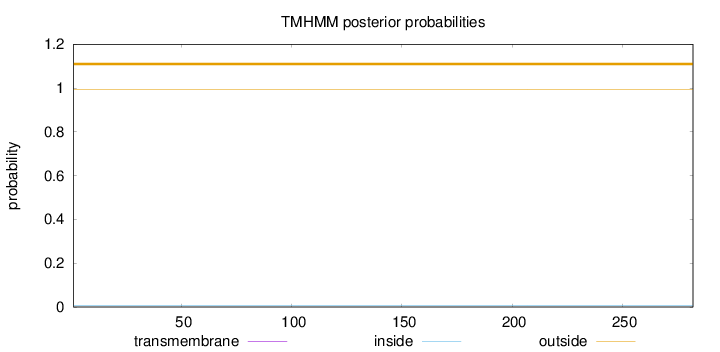
Topology
Subcellular location
Cytoplasm
Length:
282
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00143
Exp number, first 60 AAs:
0.00033
Total prob of N-in:
0.00772
outside
1 - 282
Population Genetic Test Statistics
Pi
183.576954
Theta
19.28562
Tajima's D
-2.02405
CLR
2.77745
CSRT
0.0126493675316234
Interpretation
Possibly Positive selection
