Pre Gene Modal
BGIBMGA003620
Annotation
PREDICTED:_kinesin-like_protein_KIF13B_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.247
Sequence
CDS
ATGATGGGCTCCCCGGGGGCTGACGAAGGCGGCATCATTCCTCGGCTTTGCGACGCCCTCTTTGAGAGGATAGCTGTACAACAGACACCGCCCACTCTCACGTACAAGGTGGAAGTATCATACATGGAGATATATAACGAGCGGGTGCATGATCTCCTAGATCCGGAGACCACGCGGAGGTCGCTACGAGTACGAGAGCATGCTGTCCTCGGTCCATACGTTGACGGGCTCTCGTTGTTGGCCGTTTCCTCGTTTAAGGATATCGACAATTTAATGACGGAAGGTAATAAGTCTCGAACAGTGGCAGCTACGAACATGAACAGTGAGTCTTCACGATCCCACGCCGTGTTCAGCGTTGTTCTGACTCAGACTCTATGTGATGCTGCTACCGGTGTCACCGGGGAGAAGGTTGCCAGATTATCCCTAGTGGATCTAGCCGGATCAGAGAGGGCCGTGAAGACTGGTGCTGTTGGAGACAGGCTCAAGGAAGGATCTAATATCAACAAATCTCTGACTACCCTGGGTCTCGTCATTTCAAAGTTGGCTGATCAATCGTCGGGCAAAACGAATAAGGACAAATTCGTACCGTACCGAGACTCCGTACTGACTTGGTTGCTGAAAGACAACCTCGGCGGCAACAGTAAAACTGTTATGGTGGCAACGATATCTCCAGCGGCCGACAATTACGAAGAAACGCTGTCGACCCTGAGGTACGCGGACCGAGCCAAGCGCATCGTGAACCACGCTGTGGTCAACGAGGATCCCAACGCTCGCATTATCAGGGAGCTCCGACAGGAGGTCGAAGCACTCAAGGAGATGCTCAAATACGCAACTGGATCACCCGTCGGAGAAGACGTCCATGAACAGCTCGCGCAAAGCGAACATCTGATGAAAGAGATGTCGCGGACCTGGGAGGAGAAGCTGGTCGAAACTGAACGAATACAAGGCGAACGTCAACACGCTTTGGAGAAGATGGGGATCAGCGTGCAAGCGAGCGGCATTAAAGTGGAGAAGAACAAATATTACCTCGTCAATTTGAACGCGGATCCGTCGCTTAATGAACTACTGGTCTACTATTTAAAGGAGAGGACCCTCGTGGGCGCGGACAGCTCGGCCGACATCCAGCTGTCTGGGCTGGGCATACAGCCGCAGCACTGCCTGCTGCTGGTGGAGGGCGGCGGGCTGCACATGGAGCCGGTGGGCGCCGCGCGCTGCTTCGTCAACGGCACGCCCGTCGTGTCGCGCGTGCGCCTCGGACACGCCGACCGGATCCTGTGGGGGAACCACCACTTCTTCAGGGCTAACTGCCCGAAGAAAGCGACCGGGGCCTCGGCCGGCGCGTCCGAGCCGCAGACGCCGGCGCAGCAGATCGACTACAACTTCGCTCGAGACGAGATCATGCTCAACGAGCTCAGCAACGACCCCATCCAGACGGCCATCTCGCGCCTCGAGCGCCAGCACGAGGAGGACAAGCAAGTGGCCTTAGAGAAACAACGGCAGGAATACGAAAGGCAATTTCAGCAGCTCAGGAATATACTGTCTCCGTCAACCCCGTACCCGCCCTACTCCTACGATCCGCTCAAATTAGGCAAGCTGACGGCGTGCACGCCCACCACGGCGGCGCGCGTGGAGCGCTGGGCGGCCGAGAGGGACGACACCTTCAAGCGCTCCCTGGGGCGCCTTAAGGCGGACATCCTGCGAGCCAACGCACTGGTGCAGGAGGCCAATTTCCTGGCCGAGGAGATGCGCAGGAACACCCGCTTCAGCGTCACCCTGCAGATACCGCCGCAGAACCTCAGCCCCAATCGCAAGCGCGGGGCTTTCGTGTCTGAGCCTGCTATAATGGTGAAGCGGCCAGGTCGGGGCGGCCAAGTGTGGTCAATGGAGAAACTCGATAACAAATTGGTCGACATGAGAGAAATGTATGACGAGTGGCGACGTCGACAACAACGAGGCGAGGAAGAACTGGAGGAGATTCAAACTGAGAAGACTGTGGATCCCTTCTACGAGTCTCAAGAGAACCATAATTTGATCGGCGTTGCCAACGTGTTCCTCGAGGCGTTATTCCATGACGTCACTCTCGACTATCACACCCCCATCATCAGCCAACAGGGAGAAGTGGCTGGAAGGTTACAAGTGGAGATAAGTCGCGTGGCAGGTCAATTTCCCCAAGATCGCATCTGTGAAGCTGCATCAGATAGCTCCGGTGACATGAGAGATGATGATGAACCCGTTGATCCTGCGTCACAACAGGTGACGATCCGCGTCGTAATAAAACAGGCGAGCGGGCTGCCGCCGTCTCTGTCGCACTTCGTGTTCTGTCAGTACTCGGTGTGGGGGGGAGGGGAGGCCGTGGTGGTGCCGCCCGCAGTGTGCGCCGAGCCGCGCCCCGCGCCGCACGTCACGCATCGCTTCGATCACGCCAGGGACTTCACTGTGCCGCTTACCGAGGAATTCATCGAGCATTGTGCAGAGGGTGCTCTGTCGATCGAAGTGTGGGGCCACCGGTCGGCCGGGTTCAGTCGCGCGCGCGGCAACTGGGAGGTGGAGCAGCAGCAGCTGGCGCAGGCGCGCTCGCTGGCCGACCGCTGGGCCGAGCTCTCCAGGAAGATCCAGCTCTGGGTGCAGATACACGAGCTCAACGAGCAGGGAGAATACGCGCCGGTCGAGGTGGTGGCGCGCGGCGACATGCTGACGGGCGGCGTGTACCAGCTGCGTCAGGGTCAGCAGCGGCGGATCGCAGTGCGCGTGAAGCCCGCACAGAACTCGGGGACGCTGCCGATCATATGTCAGCACATTGTCAGCGTCGAGGTGGGCTCGGTCACGGTCAGATCCCGCTTACAAAAACCTTTGGATTCATATCAGGAAGAGGACCTGGCCACGCTGCGGGAACGTTGGAGCGACGCTCTGGCCCGGCGCAGACAACACTTGCACGCACAGCTACAGAAATTGGTAAACATGTCGCCTTCGATGAAGAGTGAACGCGACATGGAACGCGAACAGTCTCTAGTCGAGCAGTGGGTGTCGCTCACCGAAGAGCGGAATGCAGTTCTAGTCCCGAGTCCTGGCAGCCCCATCCCGGGCGCTCCGGCCGCCTGGACGCCTCCGCCCGGCATGGAGCAACACATCCCCGTGCTGTTCCTTGATCTCAACGCTGACGACCTGACCACGCACCCGAGCGGCGAGGAGGTGAGCATGGCGGGGCTGAACTCCATCCTCCCCAAGGAGCTCGGCCACAAGTTCTACGAGCTGCAGATCCTGCACCACCACGACAAGGACGTCTCCGCCATCGCGTCCTGGGACTCCTCCATACACGACTCGCAGTACCTCAACAGGGTCACTGAAGCCAACGAACGCGTGTACCTGATACTAAAGACGACGGTGCGCCTCTCCCACCCGGCCCCCATGGATCTCATACTCAGGAAACGATTAGGTCTAAACATTTACAAACGACAGAGCCTCACCGACAGGATACGGAAGAAAATTGTCAGAACCGATCAGCTAACCCAGACCGGTGTTATGTACGAGGTCGTGTCGAACATACCGAAGGCCTCCGAAGAGTTGGAAGAGCGCGAGAGTCTGGCGCAGATCGCGGCCACCGGCGAGGAGGCCAGCGCCGCCGACGGAGAAACCTACATCGAGAAGTACACCCGTGGAGTGAGTGCCGTGGAGAGCATCCTGACGCTGGACCGGCTCCGCCAGAGCGTGGCCGTCAAGGAACTGGAGCAGGCGCGAGGACAGCCCATCATCATGAGGAAGACGGCTTCCGTGCCCAACTTCTCACAGCTGTCGCAGGCTCTCGAGACCGCCGCCAAGAACGGAACCAATATAATGCGCTTGGATTCGTCGTTCGAGTCGCTGACGGGGCTCGGGTCGGCGCGGTCGGGCAGCGTGGCGGACCTGGCGGACGAGAGCCCGCGGCGCGCGGCGCACCGGCACGCCGACCCCGACGCGCCGACGCCCACCAAGCCGTTCGGGCTCGCGCGGCCAACATTCCTAAACTTGAATTTGAACCTGAACTCACTGCGACTGCAGACGCCGAACAAGTCAGCATCACCGGCCAGCGGCAAGCTTGGGTTACGCATGACGACGCTGCACGAGGAGCCGGGCAGGAGGAACGACGAGGACTCTCATTCCGAACCGGAATCAGCTCCTTTGGATGAACTCACCACGCCTCGATCATCGCGCACGCGGTCTCGCGCGTCGTCCCGCGGCTTCCTGCCGACGTCGAAGACCCTCGACTCTCTGCACGAGCTGGCCTGCGAGCGAGCGCCCAACAAGAACTCTCCGTCCATATCGTCCAGTGGCTACGGTTCACAAGCGGTGTCATCCACCAATCTAACAAACGACGACACTCTGTCCATCCGGAGCATGTCTGTCGATGACACTCCAGACTTTGACAAATCCATGGATTACACAAACTTGAGCCGAACGAAGAACTCTATGGTCGGACTTCGCAATGAGATCACCGAGCTCAGAAACGACATAAATGATCTAAAAAACGACATAGTCGAGTCCAAGAATGAACTCGAAGATGCGACTTTTGCCAAGACTCCAGTGCTGACTCCCGGCTCGAGGAACCGGATCAATCCGTTCCTGAGGGATTGCGAGGATGCCGTTAAAGAAGAGAGTAAGGACCAGCTTGTGGATCCATTGGGATCCATACCAGTTGAGAACGCAGTGCTGTCTAGTCAAACGAAGAGCCCAGAACCGGTACAAGTCAACGGTGAAGTGTCTCATGATAATTCTTTCGGTGAAGCAGAAACAGAGTCTGTGAGCTCGGAGCCGTCGAGTCACGAAGCGGAGCGGAGCCGCGAGGCGTCGTCGGTGGGGGAGAGCGACTCGGCCTCGCTGGGCGCGGACGGCCCCGTGGTGAAGCCGCAGCTGCCCGCCGGCAAGCTGGTGCGGCGCCGCACGGGGGGCGGGCGGGCGGCGCGCGGGCCCGCCCTGCGCCCGCGCTCCATGCACGAGACCTCGTCCGCCTCCATGCTGTCTAGCTCGCCGGCTTCCTCTACGGAAAGAATTGGAGAGGACGACACGTCGGAGGGCACGATCCAGCCGTGGATGACGGTGGGTGAGAGCGTCCAGCTCCGCCTGAGCACAAGCACCGGCCTCATCGCGTACACCGGGCCCACGCACTTCGCGCCCGGCACCTGGGTCGGGGTGCAACTCGACGCTCCAACGGGCAAAAACGACGGTTCGGTGGGCGGCACCCGGTACTTCGAGTGTCGGCCGCGGCACGGCATCTTCGTGCGCCCCGACAAGCTGGTGCAGGACCGCCGCGGCCGCTCCGCGCGCCAGTACGCGCACCTCGCCCGCGCCTCCAGCAAGGGGGAAGGCCTACATAACCTGCACAGGTCAAGGAGTCGCGGCGACAGTATAAACACAATCGGTAAAACCAGAAGCAAATAG
Protein
MMGSPGADEGGIIPRLCDALFERIAVQQTPPTLTYKVEVSYMEIYNERVHDLLDPETTRRSLRVREHAVLGPYVDGLSLLAVSSFKDIDNLMTEGNKSRTVAATNMNSESSRSHAVFSVVLTQTLCDAATGVTGEKVARLSLVDLAGSERAVKTGAVGDRLKEGSNINKSLTTLGLVISKLADQSSGKTNKDKFVPYRDSVLTWLLKDNLGGNSKTVMVATISPAADNYEETLSTLRYADRAKRIVNHAVVNEDPNARIIRELRQEVEALKEMLKYATGSPVGEDVHEQLAQSEHLMKEMSRTWEEKLVETERIQGERQHALEKMGISVQASGIKVEKNKYYLVNLNADPSLNELLVYYLKERTLVGADSSADIQLSGLGIQPQHCLLLVEGGGLHMEPVGAARCFVNGTPVVSRVRLGHADRILWGNHHFFRANCPKKATGASAGASEPQTPAQQIDYNFARDEIMLNELSNDPIQTAISRLERQHEEDKQVALEKQRQEYERQFQQLRNILSPSTPYPPYSYDPLKLGKLTACTPTTAARVERWAAERDDTFKRSLGRLKADILRANALVQEANFLAEEMRRNTRFSVTLQIPPQNLSPNRKRGAFVSEPAIMVKRPGRGGQVWSMEKLDNKLVDMREMYDEWRRRQQRGEEELEEIQTEKTVDPFYESQENHNLIGVANVFLEALFHDVTLDYHTPIISQQGEVAGRLQVEISRVAGQFPQDRICEAASDSSGDMRDDDEPVDPASQQVTIRVVIKQASGLPPSLSHFVFCQYSVWGGGEAVVVPPAVCAEPRPAPHVTHRFDHARDFTVPLTEEFIEHCAEGALSIEVWGHRSAGFSRARGNWEVEQQQLAQARSLADRWAELSRKIQLWVQIHELNEQGEYAPVEVVARGDMLTGGVYQLRQGQQRRIAVRVKPAQNSGTLPIICQHIVSVEVGSVTVRSRLQKPLDSYQEEDLATLRERWSDALARRRQHLHAQLQKLVNMSPSMKSERDMEREQSLVEQWVSLTEERNAVLVPSPGSPIPGAPAAWTPPPGMEQHIPVLFLDLNADDLTTHPSGEEVSMAGLNSILPKELGHKFYELQILHHHDKDVSAIASWDSSIHDSQYLNRVTEANERVYLILKTTVRLSHPAPMDLILRKRLGLNIYKRQSLTDRIRKKIVRTDQLTQTGVMYEVVSNIPKASEELEERESLAQIAATGEEASAADGETYIEKYTRGVSAVESILTLDRLRQSVAVKELEQARGQPIIMRKTASVPNFSQLSQALETAAKNGTNIMRLDSSFESLTGLGSARSGSVADLADESPRRAAHRHADPDAPTPTKPFGLARPTFLNLNLNLNSLRLQTPNKSASPASGKLGLRMTTLHEEPGRRNDEDSHSEPESAPLDELTTPRSSRTRSRASSRGFLPTSKTLDSLHELACERAPNKNSPSISSSGYGSQAVSSTNLTNDDTLSIRSMSVDDTPDFDKSMDYTNLSRTKNSMVGLRNEITELRNDINDLKNDIVESKNELEDATFAKTPVLTPGSRNRINPFLRDCEDAVKEESKDQLVDPLGSIPVENAVLSSQTKSPEPVQVNGEVSHDNSFGEAETESVSSEPSSHEAERSREASSVGESDSASLGADGPVVKPQLPAGKLVRRRTGGGRAARGPALRPRSMHETSSASMLSSSPASSTERIGEDDTSEGTIQPWMTVGESVQLRLSTSTGLIAYTGPTHFAPGTWVGVQLDAPTGKNDGSVGGTRYFECRPRHGIFVRPDKLVQDRRGRSARQYAHLARASSKGEGLHNLHRSRSRGDSINTIGKTRSK
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
A0A2H1WP10
A0A194PP51
A0A1E1VY35
A0A1E1W0F9
A0A212FBU9
A0A151IIB9
+ More
A0A0P9AEB6 A0A0P8XNY0 A0A0N8P072 A0A0P9BXX3 A0A0P8XP04 A0A0P8Y3R4 A0A2P8YHN1 A0A0P8XNU8 A0A0P8YBA0 B3MEY4 A0A1Y1KRN4 A0A1Y1KNU4 E2C5X9 A0A088AVW8 E2ARY8 A0A1W4W0C2 A0A1W4W0B7 A0A1Y1KTQ0 A0A0R1DWQ6 A0A0R1DYD8 A0A1Y1KNU5 A0A1W4W0B9 A0A1W4WCE2 A0A0J9RDF1 A0A0B4KG24 A0A0Q5VYH6 A0A0R1DYD6 A0A0Q5VLK6 A0A1W4WB23 A0A0J9RDK2 A0A0J9REL2 A0A1B6EBU6 A0A0Q5W175 A0A0J9U2R5 B4P7C4 A0A0R1DYR6 A0A1W4WCE7 A0A1W4WC02 A0A0J9RDE6 A0A0Q5VNK9 B3NQC1 A0A0J9RDJ5 A0A1W4W0C5 A0A195B7F0 A0A0Q9WXV6 A0A0J9U2R2 A0A0B4KFR5 A4UZI0 A0A1W4WB28 A0A3L8D648 A1ZA18 A0A195DWJ4 A0A0L7QSR5 A0A0B4LGK5 F4WZT9 O01349 B4QGV6 W8AWR0 A0A0Q9X7K1 A0A026VY99 A0A1B6C9W7 A0A0K8VNS8 W8AHV3 A0A3B0JQZ3 A0A0K8VYY3 A0A0A1XLP7 A0A0Q9W7P5 N6WC38 A0A0K8W954 B4NMT9 A0A3B0J3A3 A0A0Q9WDZ4 B4LP34 A0A3B0J398 A0A0T6BFU7 A0A3B0J2S5 B4J5N9 A0A139WNS3 A0A0M5JAA6 A0A1I8P0J2 A0A195FFP0 A0A1I8P0I8 A0A1B0DN90 A0A1I8P0E7 A0A0C9PHU7 A0A1I8P0H8 Q17KS2 D6W757 A0A1I8P0J7 W4VRD3 A0A0C9RH69 A0A1I8P0I3 A0A0C9PHU2
A0A0P9AEB6 A0A0P8XNY0 A0A0N8P072 A0A0P9BXX3 A0A0P8XP04 A0A0P8Y3R4 A0A2P8YHN1 A0A0P8XNU8 A0A0P8YBA0 B3MEY4 A0A1Y1KRN4 A0A1Y1KNU4 E2C5X9 A0A088AVW8 E2ARY8 A0A1W4W0C2 A0A1W4W0B7 A0A1Y1KTQ0 A0A0R1DWQ6 A0A0R1DYD8 A0A1Y1KNU5 A0A1W4W0B9 A0A1W4WCE2 A0A0J9RDF1 A0A0B4KG24 A0A0Q5VYH6 A0A0R1DYD6 A0A0Q5VLK6 A0A1W4WB23 A0A0J9RDK2 A0A0J9REL2 A0A1B6EBU6 A0A0Q5W175 A0A0J9U2R5 B4P7C4 A0A0R1DYR6 A0A1W4WCE7 A0A1W4WC02 A0A0J9RDE6 A0A0Q5VNK9 B3NQC1 A0A0J9RDJ5 A0A1W4W0C5 A0A195B7F0 A0A0Q9WXV6 A0A0J9U2R2 A0A0B4KFR5 A4UZI0 A0A1W4WB28 A0A3L8D648 A1ZA18 A0A195DWJ4 A0A0L7QSR5 A0A0B4LGK5 F4WZT9 O01349 B4QGV6 W8AWR0 A0A0Q9X7K1 A0A026VY99 A0A1B6C9W7 A0A0K8VNS8 W8AHV3 A0A3B0JQZ3 A0A0K8VYY3 A0A0A1XLP7 A0A0Q9W7P5 N6WC38 A0A0K8W954 B4NMT9 A0A3B0J3A3 A0A0Q9WDZ4 B4LP34 A0A3B0J398 A0A0T6BFU7 A0A3B0J2S5 B4J5N9 A0A139WNS3 A0A0M5JAA6 A0A1I8P0J2 A0A195FFP0 A0A1I8P0I8 A0A1B0DN90 A0A1I8P0E7 A0A0C9PHU7 A0A1I8P0H8 Q17KS2 D6W757 A0A1I8P0J7 W4VRD3 A0A0C9RH69 A0A1I8P0I3 A0A0C9PHU2
Pubmed
EMBL
ODYU01009972
SOQ54747.1
KQ459597
KPI95087.1
GDQN01011460
JAT79594.1
+ More
GDQN01010574 JAT80480.1 AGBW02009273 OWR51203.1 KQ977492 KYN02300.1 CH902619 KPU76318.1 KPU76319.1 KPU76316.1 KPU76322.1 KPU76320.1 KPU76315.1 PYGN01000589 PSN43743.1 KPU76317.1 KPU76321.1 EDV36605.2 GEZM01077996 JAV63071.1 GEZM01077995 JAV63072.1 GL452874 EFN76653.1 GL442209 EFN63762.1 GEZM01077993 JAV63075.1 CM000158 KRK00307.1 KRK00306.1 GEZM01077994 JAV63073.1 CM002911 KMY94007.1 AE013599 AGB93531.1 CH954179 KQS62542.1 KRK00305.1 KQS62541.1 KMY94006.1 KMY94004.1 GEDC01001893 JAS35405.1 KQS62543.1 KMY94005.1 EDW92069.2 KRK00308.1 KMY94002.1 KQS62540.1 EDV55897.2 KMY94001.1 KQ976574 KYM80175.1 CH964282 KRG00265.1 KMY94000.1 AGB93532.1 AAF58129.2 QOIP01000012 RLU15880.1 AAM70975.1 KQ980204 KYN17216.1 KQ414755 KOC61693.1 AHN56259.1 GL888480 EGI60364.1 U81788 AAB50404.1 CM000362 EDX07219.1 KMY94003.1 GAMC01021310 GAMC01021304 JAB85251.1 CH933808 KRG04236.1 KK107770 EZA47844.1 GEDC01027016 JAS10282.1 GDHF01011777 JAI40537.1 GAMC01021308 GAMC01021306 JAB85247.1 OUUW01000001 SPP75786.1 GDHF01008248 JAI44066.1 GBXI01002371 JAD11921.1 CH940648 KRF80502.1 CM000071 ENO01734.2 GDHF01004740 JAI47574.1 EDW85678.2 SPP75785.1 KRF80503.1 EDW62228.2 SPP75787.1 LJIG01000732 KRT86208.1 SPP75784.1 CH916367 EDW01815.1 KQ971307 KYB29658.1 CP012524 ALC41981.1 KQ981625 KYN39171.1 AJVK01007438 AJVK01007439 AJVK01007440 GBYB01000453 JAG70220.1 CH477221 EAT47335.1 EFA11512.2 GANO01003561 JAB56310.1 GBYB01012557 JAG82324.1 GBYB01000448 JAG70215.1
GDQN01010574 JAT80480.1 AGBW02009273 OWR51203.1 KQ977492 KYN02300.1 CH902619 KPU76318.1 KPU76319.1 KPU76316.1 KPU76322.1 KPU76320.1 KPU76315.1 PYGN01000589 PSN43743.1 KPU76317.1 KPU76321.1 EDV36605.2 GEZM01077996 JAV63071.1 GEZM01077995 JAV63072.1 GL452874 EFN76653.1 GL442209 EFN63762.1 GEZM01077993 JAV63075.1 CM000158 KRK00307.1 KRK00306.1 GEZM01077994 JAV63073.1 CM002911 KMY94007.1 AE013599 AGB93531.1 CH954179 KQS62542.1 KRK00305.1 KQS62541.1 KMY94006.1 KMY94004.1 GEDC01001893 JAS35405.1 KQS62543.1 KMY94005.1 EDW92069.2 KRK00308.1 KMY94002.1 KQS62540.1 EDV55897.2 KMY94001.1 KQ976574 KYM80175.1 CH964282 KRG00265.1 KMY94000.1 AGB93532.1 AAF58129.2 QOIP01000012 RLU15880.1 AAM70975.1 KQ980204 KYN17216.1 KQ414755 KOC61693.1 AHN56259.1 GL888480 EGI60364.1 U81788 AAB50404.1 CM000362 EDX07219.1 KMY94003.1 GAMC01021310 GAMC01021304 JAB85251.1 CH933808 KRG04236.1 KK107770 EZA47844.1 GEDC01027016 JAS10282.1 GDHF01011777 JAI40537.1 GAMC01021308 GAMC01021306 JAB85247.1 OUUW01000001 SPP75786.1 GDHF01008248 JAI44066.1 GBXI01002371 JAD11921.1 CH940648 KRF80502.1 CM000071 ENO01734.2 GDHF01004740 JAI47574.1 EDW85678.2 SPP75785.1 KRF80503.1 EDW62228.2 SPP75787.1 LJIG01000732 KRT86208.1 SPP75784.1 CH916367 EDW01815.1 KQ971307 KYB29658.1 CP012524 ALC41981.1 KQ981625 KYN39171.1 AJVK01007438 AJVK01007439 AJVK01007440 GBYB01000453 JAG70220.1 CH477221 EAT47335.1 EFA11512.2 GANO01003561 JAB56310.1 GBYB01012557 JAG82324.1 GBYB01000448 JAG70215.1
Proteomes
UP000053268
UP000007151
UP000078542
UP000007801
UP000245037
UP000008237
+ More
UP000005203 UP000000311 UP000192221 UP000002282 UP000000803 UP000008711 UP000078540 UP000007798 UP000279307 UP000078492 UP000053825 UP000007755 UP000000304 UP000009192 UP000053097 UP000268350 UP000008792 UP000001819 UP000001070 UP000007266 UP000092553 UP000095300 UP000078541 UP000092462 UP000008820
UP000005203 UP000000311 UP000192221 UP000002282 UP000000803 UP000008711 UP000078540 UP000007798 UP000279307 UP000078492 UP000053825 UP000007755 UP000000304 UP000009192 UP000053097 UP000268350 UP000008792 UP000001819 UP000001070 UP000007266 UP000092553 UP000095300 UP000078541 UP000092462 UP000008820
Pfam
Interpro
IPR027640
Kinesin-like_fam
+ More
IPR022140 Kinesin-like_KIF1-typ
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR032405 Kinesin_assoc
IPR000938 CAP-Gly_domain
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR022164 Kinesin-like
IPR036859 CAP-Gly_dom_sf
IPR000253 FHA_dom
IPR008984 SMAD_FHA_dom_sf
IPR032923 KIF13A
IPR022140 Kinesin-like_KIF1-typ
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR032405 Kinesin_assoc
IPR000938 CAP-Gly_domain
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR022164 Kinesin-like
IPR036859 CAP-Gly_dom_sf
IPR000253 FHA_dom
IPR008984 SMAD_FHA_dom_sf
IPR032923 KIF13A
Gene 3D
CDD
ProteinModelPortal
A0A2H1WP10
A0A194PP51
A0A1E1VY35
A0A1E1W0F9
A0A212FBU9
A0A151IIB9
+ More
A0A0P9AEB6 A0A0P8XNY0 A0A0N8P072 A0A0P9BXX3 A0A0P8XP04 A0A0P8Y3R4 A0A2P8YHN1 A0A0P8XNU8 A0A0P8YBA0 B3MEY4 A0A1Y1KRN4 A0A1Y1KNU4 E2C5X9 A0A088AVW8 E2ARY8 A0A1W4W0C2 A0A1W4W0B7 A0A1Y1KTQ0 A0A0R1DWQ6 A0A0R1DYD8 A0A1Y1KNU5 A0A1W4W0B9 A0A1W4WCE2 A0A0J9RDF1 A0A0B4KG24 A0A0Q5VYH6 A0A0R1DYD6 A0A0Q5VLK6 A0A1W4WB23 A0A0J9RDK2 A0A0J9REL2 A0A1B6EBU6 A0A0Q5W175 A0A0J9U2R5 B4P7C4 A0A0R1DYR6 A0A1W4WCE7 A0A1W4WC02 A0A0J9RDE6 A0A0Q5VNK9 B3NQC1 A0A0J9RDJ5 A0A1W4W0C5 A0A195B7F0 A0A0Q9WXV6 A0A0J9U2R2 A0A0B4KFR5 A4UZI0 A0A1W4WB28 A0A3L8D648 A1ZA18 A0A195DWJ4 A0A0L7QSR5 A0A0B4LGK5 F4WZT9 O01349 B4QGV6 W8AWR0 A0A0Q9X7K1 A0A026VY99 A0A1B6C9W7 A0A0K8VNS8 W8AHV3 A0A3B0JQZ3 A0A0K8VYY3 A0A0A1XLP7 A0A0Q9W7P5 N6WC38 A0A0K8W954 B4NMT9 A0A3B0J3A3 A0A0Q9WDZ4 B4LP34 A0A3B0J398 A0A0T6BFU7 A0A3B0J2S5 B4J5N9 A0A139WNS3 A0A0M5JAA6 A0A1I8P0J2 A0A195FFP0 A0A1I8P0I8 A0A1B0DN90 A0A1I8P0E7 A0A0C9PHU7 A0A1I8P0H8 Q17KS2 D6W757 A0A1I8P0J7 W4VRD3 A0A0C9RH69 A0A1I8P0I3 A0A0C9PHU2
A0A0P9AEB6 A0A0P8XNY0 A0A0N8P072 A0A0P9BXX3 A0A0P8XP04 A0A0P8Y3R4 A0A2P8YHN1 A0A0P8XNU8 A0A0P8YBA0 B3MEY4 A0A1Y1KRN4 A0A1Y1KNU4 E2C5X9 A0A088AVW8 E2ARY8 A0A1W4W0C2 A0A1W4W0B7 A0A1Y1KTQ0 A0A0R1DWQ6 A0A0R1DYD8 A0A1Y1KNU5 A0A1W4W0B9 A0A1W4WCE2 A0A0J9RDF1 A0A0B4KG24 A0A0Q5VYH6 A0A0R1DYD6 A0A0Q5VLK6 A0A1W4WB23 A0A0J9RDK2 A0A0J9REL2 A0A1B6EBU6 A0A0Q5W175 A0A0J9U2R5 B4P7C4 A0A0R1DYR6 A0A1W4WCE7 A0A1W4WC02 A0A0J9RDE6 A0A0Q5VNK9 B3NQC1 A0A0J9RDJ5 A0A1W4W0C5 A0A195B7F0 A0A0Q9WXV6 A0A0J9U2R2 A0A0B4KFR5 A4UZI0 A0A1W4WB28 A0A3L8D648 A1ZA18 A0A195DWJ4 A0A0L7QSR5 A0A0B4LGK5 F4WZT9 O01349 B4QGV6 W8AWR0 A0A0Q9X7K1 A0A026VY99 A0A1B6C9W7 A0A0K8VNS8 W8AHV3 A0A3B0JQZ3 A0A0K8VYY3 A0A0A1XLP7 A0A0Q9W7P5 N6WC38 A0A0K8W954 B4NMT9 A0A3B0J3A3 A0A0Q9WDZ4 B4LP34 A0A3B0J398 A0A0T6BFU7 A0A3B0J2S5 B4J5N9 A0A139WNS3 A0A0M5JAA6 A0A1I8P0J2 A0A195FFP0 A0A1I8P0I8 A0A1B0DN90 A0A1I8P0E7 A0A0C9PHU7 A0A1I8P0H8 Q17KS2 D6W757 A0A1I8P0J7 W4VRD3 A0A0C9RH69 A0A1I8P0I3 A0A0C9PHU2
PDB
6A20
E-value=1.00077e-120,
Score=1115
Ontologies
GO
PANTHER
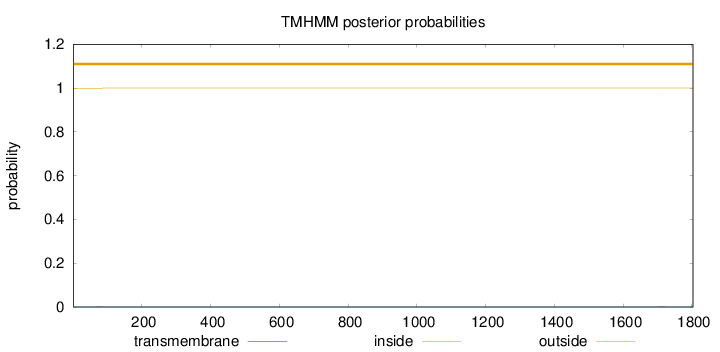
Topology
Length:
1803
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01578
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00053
outside
1 - 1803
Population Genetic Test Statistics
Pi
238.786216
Theta
184.973115
Tajima's D
0.62136
CLR
0.703927
CSRT
0.546822658867057
Interpretation
Uncertain
