Gene
KWMTBOMO02981
Pre Gene Modal
BGIBMGA003622
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_dynein_beta_chain?_ciliary-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.123
Sequence
CDS
ATGTGCGGCAAGGAAGCTGAATTCAAAGCAATTTTATTCTCGTTGTGCTACTTCCACGCTGTGGTTGCTGAGAGGCGCAAGTTCGGTCCTCAAGGATGGAATAAAATTTATCCATTTAACGTTGGTGATCTCACCATTAGCGTATTCGTTCTGTACAACTATTTGGAAGCAAATTCAAGGGTGCCCTGGGAAGATTTGAGATATCTCTTCGGAGAGATAATGTACGGAGGTCATATAACTGATGATTGGGACAGAAGGCTCTGTAACGCATACCTGGAAGAATTAATGCAGCCGGATCTGGTTGATGGAGAGCTGCAACTCGCCCCAGGTTTCCCTGCTCCACCAAACACAGATTACAGCGGCTACCATCAATACATAGAAGACGCCATGCCGGCTGAATCTCCGTACCTTTACGGGCTACATCCAAATGCTGAAATAGGTTTTCTGACGACTACCTCAGAAAATTTGTTTAGAACGATATTCGAGATGCAACCAAGAGATGCTCAGGCTTCTGGTGCCACTACTGTGACCAGAGAGGACAAGGTTTGTTTCATGGTTTTTTAG
Protein
MCGKEAEFKAILFSLCYFHAVVAERRKFGPQGWNKIYPFNVGDLTISVFVLYNYLEANSRVPWEDLRYLFGEIMYGGHITDDWDRRLCNAYLEELMQPDLVDGELQLAPGFPAPPNTDYSGYHQYIEDAMPAESPYLYGLHPNAEIGFLTTTSENLFRTIFEMQPRDAQASGATTVTREDKVCFMVF
Summary
Uniprot
H9J285
A0A2H1X2P9
A0A2A4JJP5
A0A0N1ID47
A0A212FLC8
A0A1B6DVH7
+ More
A0A2J7PBF2 A0A2J7PBE9 A0A2J7RTB7 A0A2J7RTB8 A0A034UZR8 A0A2J7RTA3 A0A2J7RTA2 A0A0A9Y300 A0A2J7RTA7 A0A2J7RT97 A0A139WPL9 D6W7L1 A0A182SWG7 A0A1B0EYP6 E0VLA6 W8CAW5 A0A1J1IHV8 A0A2M4DP19 A0A232F8L3 A0A182V866 B0WUU8 A0A182IVR3 A0A1B6GIG4 A0A182YIP1 W5J456 A0A084VZI7 A0A182MNY4 A0A182LNA0 A0A2M4A0B6 A0A182FM39 A0A182NDL4 A0A1S4H4T4 A0A182W3T1 A0A182WR89 A0A182JRE6 A0A182PVS5 A0A182HX97 A0A182QB97 Q5TUD8 A0A182RBC6 A0A182TS04 A0A0M9AA00 A0A1S4FKM6 Q16XH8 A0A154PPG4 A0A182GMZ9 A0A336KXF6 A0A1W4X3T6 U4UMC9 A0A0R3NKH1 Q293J4 A0A336M9T6 A0A2S2QK24 N6U0M0 A0A0L7QYM1 E9IZF0 A0A1A9WGW2 A0A310S6L6 A0A1I8PWQ2 A0A195CXR3 A0A151JQJ9 A0A151JXC9 A0A1B0G9S0 A0A3B0JSI1 A0A1A9UNH8 A0A1A9XQ49 A0A1A9ZDG9 A0A0R1E127 A0A0B4KHJ4 A0A0R1E5P0 Q9VDG0 A0A1I8M6W5 A0A0R1E1G1 B3P2Z8 A0A0R1E125 B4IKM6 A0A1B0BD95 A0A195AUI3 A0A1I8M6W0 B4PQ99 B4R1N9 A0A0P8Y0J0 A0A1W4VZS9 A0A1W4VLP2 B3M2X7 B4M4V0 A0A158P169 A0A0L0BTW6 A0A0J7P079 B4JSY1 E2BE63 A0A151WVZ6 B4KE24 E2AYZ0
A0A2J7PBF2 A0A2J7PBE9 A0A2J7RTB7 A0A2J7RTB8 A0A034UZR8 A0A2J7RTA3 A0A2J7RTA2 A0A0A9Y300 A0A2J7RTA7 A0A2J7RT97 A0A139WPL9 D6W7L1 A0A182SWG7 A0A1B0EYP6 E0VLA6 W8CAW5 A0A1J1IHV8 A0A2M4DP19 A0A232F8L3 A0A182V866 B0WUU8 A0A182IVR3 A0A1B6GIG4 A0A182YIP1 W5J456 A0A084VZI7 A0A182MNY4 A0A182LNA0 A0A2M4A0B6 A0A182FM39 A0A182NDL4 A0A1S4H4T4 A0A182W3T1 A0A182WR89 A0A182JRE6 A0A182PVS5 A0A182HX97 A0A182QB97 Q5TUD8 A0A182RBC6 A0A182TS04 A0A0M9AA00 A0A1S4FKM6 Q16XH8 A0A154PPG4 A0A182GMZ9 A0A336KXF6 A0A1W4X3T6 U4UMC9 A0A0R3NKH1 Q293J4 A0A336M9T6 A0A2S2QK24 N6U0M0 A0A0L7QYM1 E9IZF0 A0A1A9WGW2 A0A310S6L6 A0A1I8PWQ2 A0A195CXR3 A0A151JQJ9 A0A151JXC9 A0A1B0G9S0 A0A3B0JSI1 A0A1A9UNH8 A0A1A9XQ49 A0A1A9ZDG9 A0A0R1E127 A0A0B4KHJ4 A0A0R1E5P0 Q9VDG0 A0A1I8M6W5 A0A0R1E1G1 B3P2Z8 A0A0R1E125 B4IKM6 A0A1B0BD95 A0A195AUI3 A0A1I8M6W0 B4PQ99 B4R1N9 A0A0P8Y0J0 A0A1W4VZS9 A0A1W4VLP2 B3M2X7 B4M4V0 A0A158P169 A0A0L0BTW6 A0A0J7P079 B4JSY1 E2BE63 A0A151WVZ6 B4KE24 E2AYZ0
Pubmed
19121390
26354079
22118469
25348373
25401762
18362917
+ More
19820115 20566863 24495485 28648823 25244985 20920257 23761445 24438588 20966253 12364791 17510324 26483478 23537049 15632085 17994087 21282665 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 21347285 26108605 20798317
19820115 20566863 24495485 28648823 25244985 20920257 23761445 24438588 20966253 12364791 17510324 26483478 23537049 15632085 17994087 21282665 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 21347285 26108605 20798317
EMBL
BABH01007454
BABH01007455
BABH01007456
BABH01007457
ODYU01012953
SOQ59508.1
+ More
NWSH01001294 PCG71814.1 KQ461187 KPJ07329.1 AGBW02007827 OWR54500.1 GEDC01007614 JAS29684.1 NEVH01027098 PNF13660.1 PNF13659.1 NEVH01000006 PNF44066.1 PNF44069.1 GAKP01023217 JAC35739.1 PNF44070.1 PNF44065.1 GBHO01018116 JAG25488.1 PNF44067.1 PNF44063.1 KQ971307 KYB29781.1 EFA11309.2 AJVK01064446 AAZO01003313 DS235271 EEB14162.1 GAMC01000989 JAC05567.1 CVRI01000047 CRK98041.1 GGFL01015101 MBW79279.1 NNAY01000755 OXU26657.1 DS232111 EDS35091.1 GECZ01007537 JAS62232.1 ADMH02002126 ETN58641.1 ATLV01018849 KE525251 KFB43381.1 AXCM01000636 GGFK01000870 MBW34191.1 AAAB01008849 APCN01005160 AXCN02001753 EAL41019.3 KQ435706 KOX80265.1 CH477538 EAT39332.1 KQ434980 KZC13060.1 JXUM01075435 KQ562886 KXJ74924.1 UFQS01001276 UFQT01001276 SSX10081.1 SSX29803.1 KB632384 ERL94272.1 CM000070 KRT01415.1 EAL29221.2 UFQS01000646 UFQT01000646 SSX05712.1 SSX26071.1 GGMS01008884 MBY78087.1 APGK01047112 APGK01047113 APGK01047114 APGK01047115 APGK01047116 APGK01047117 APGK01047118 KB741077 ENN74141.1 KQ414688 KOC63714.1 GL767121 EFZ14108.1 KQ767877 OAD53287.1 KQ977141 KYN05431.1 KQ978644 KYN29472.1 KQ981572 KYN39778.1 CCAG010011423 OUUW01000007 SPP83382.1 CM000160 KRK03003.1 AE014297 AGB96173.1 KRK03002.1 AAF55834.3 KRK03001.1 CH954181 EDV48312.1 KRK03004.1 CH480853 EDW51630.1 JXJN01012383 KQ976738 KYM75717.1 EDW96208.2 CM000364 EDX12148.1 CH902617 KPU80296.1 EDV43507.1 CH940652 EDW59661.2 ADTU01005848 ADTU01005849 JRES01001345 KNC23525.1 LBMM01000571 KMQ98045.1 CH916373 EDV94871.1 GL447755 EFN86050.1 KQ982691 KYQ52109.1 CH933806 EDW16045.2 GL444028 EFN61360.1
NWSH01001294 PCG71814.1 KQ461187 KPJ07329.1 AGBW02007827 OWR54500.1 GEDC01007614 JAS29684.1 NEVH01027098 PNF13660.1 PNF13659.1 NEVH01000006 PNF44066.1 PNF44069.1 GAKP01023217 JAC35739.1 PNF44070.1 PNF44065.1 GBHO01018116 JAG25488.1 PNF44067.1 PNF44063.1 KQ971307 KYB29781.1 EFA11309.2 AJVK01064446 AAZO01003313 DS235271 EEB14162.1 GAMC01000989 JAC05567.1 CVRI01000047 CRK98041.1 GGFL01015101 MBW79279.1 NNAY01000755 OXU26657.1 DS232111 EDS35091.1 GECZ01007537 JAS62232.1 ADMH02002126 ETN58641.1 ATLV01018849 KE525251 KFB43381.1 AXCM01000636 GGFK01000870 MBW34191.1 AAAB01008849 APCN01005160 AXCN02001753 EAL41019.3 KQ435706 KOX80265.1 CH477538 EAT39332.1 KQ434980 KZC13060.1 JXUM01075435 KQ562886 KXJ74924.1 UFQS01001276 UFQT01001276 SSX10081.1 SSX29803.1 KB632384 ERL94272.1 CM000070 KRT01415.1 EAL29221.2 UFQS01000646 UFQT01000646 SSX05712.1 SSX26071.1 GGMS01008884 MBY78087.1 APGK01047112 APGK01047113 APGK01047114 APGK01047115 APGK01047116 APGK01047117 APGK01047118 KB741077 ENN74141.1 KQ414688 KOC63714.1 GL767121 EFZ14108.1 KQ767877 OAD53287.1 KQ977141 KYN05431.1 KQ978644 KYN29472.1 KQ981572 KYN39778.1 CCAG010011423 OUUW01000007 SPP83382.1 CM000160 KRK03003.1 AE014297 AGB96173.1 KRK03002.1 AAF55834.3 KRK03001.1 CH954181 EDV48312.1 KRK03004.1 CH480853 EDW51630.1 JXJN01012383 KQ976738 KYM75717.1 EDW96208.2 CM000364 EDX12148.1 CH902617 KPU80296.1 EDV43507.1 CH940652 EDW59661.2 ADTU01005848 ADTU01005849 JRES01001345 KNC23525.1 LBMM01000571 KMQ98045.1 CH916373 EDV94871.1 GL447755 EFN86050.1 KQ982691 KYQ52109.1 CH933806 EDW16045.2 GL444028 EFN61360.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000235965
UP000007266
+ More
UP000075901 UP000092462 UP000009046 UP000183832 UP000215335 UP000075903 UP000002320 UP000075880 UP000076408 UP000000673 UP000030765 UP000075883 UP000075882 UP000069272 UP000075884 UP000075920 UP000076407 UP000075881 UP000075885 UP000075840 UP000075886 UP000007062 UP000075900 UP000075902 UP000053105 UP000008820 UP000076502 UP000069940 UP000249989 UP000192223 UP000030742 UP000001819 UP000019118 UP000053825 UP000091820 UP000095300 UP000078542 UP000078492 UP000078541 UP000092444 UP000268350 UP000078200 UP000092443 UP000092445 UP000002282 UP000000803 UP000095301 UP000008711 UP000001292 UP000092460 UP000078540 UP000000304 UP000007801 UP000192221 UP000008792 UP000005205 UP000037069 UP000036403 UP000001070 UP000008237 UP000075809 UP000009192 UP000000311
UP000075901 UP000092462 UP000009046 UP000183832 UP000215335 UP000075903 UP000002320 UP000075880 UP000076408 UP000000673 UP000030765 UP000075883 UP000075882 UP000069272 UP000075884 UP000075920 UP000076407 UP000075881 UP000075885 UP000075840 UP000075886 UP000007062 UP000075900 UP000075902 UP000053105 UP000008820 UP000076502 UP000069940 UP000249989 UP000192223 UP000030742 UP000001819 UP000019118 UP000053825 UP000091820 UP000095300 UP000078542 UP000078492 UP000078541 UP000092444 UP000268350 UP000078200 UP000092443 UP000092445 UP000002282 UP000000803 UP000095301 UP000008711 UP000001292 UP000092460 UP000078540 UP000000304 UP000007801 UP000192221 UP000008792 UP000005205 UP000037069 UP000036403 UP000001070 UP000008237 UP000075809 UP000009192 UP000000311
Pfam
Interpro
IPR042228
Dynein_2_C
+ More
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR035699 AAA_6
IPR013594 Dynein_heavy_dom-1
IPR026983 DHC_fam
IPR042219 AAA_lid_11_sf
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR027417 P-loop_NTPase
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR042222 Dynein_2_N
IPR009069 Cys_alpha_HP_mot_SF
IPR011704 ATPase_dyneun-rel_AAA
IPR036084 Ser_inhib-like_sf
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR035699 AAA_6
IPR013594 Dynein_heavy_dom-1
IPR026983 DHC_fam
IPR042219 AAA_lid_11_sf
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR027417 P-loop_NTPase
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR042222 Dynein_2_N
IPR009069 Cys_alpha_HP_mot_SF
IPR011704 ATPase_dyneun-rel_AAA
IPR036084 Ser_inhib-like_sf
Gene 3D
ProteinModelPortal
H9J285
A0A2H1X2P9
A0A2A4JJP5
A0A0N1ID47
A0A212FLC8
A0A1B6DVH7
+ More
A0A2J7PBF2 A0A2J7PBE9 A0A2J7RTB7 A0A2J7RTB8 A0A034UZR8 A0A2J7RTA3 A0A2J7RTA2 A0A0A9Y300 A0A2J7RTA7 A0A2J7RT97 A0A139WPL9 D6W7L1 A0A182SWG7 A0A1B0EYP6 E0VLA6 W8CAW5 A0A1J1IHV8 A0A2M4DP19 A0A232F8L3 A0A182V866 B0WUU8 A0A182IVR3 A0A1B6GIG4 A0A182YIP1 W5J456 A0A084VZI7 A0A182MNY4 A0A182LNA0 A0A2M4A0B6 A0A182FM39 A0A182NDL4 A0A1S4H4T4 A0A182W3T1 A0A182WR89 A0A182JRE6 A0A182PVS5 A0A182HX97 A0A182QB97 Q5TUD8 A0A182RBC6 A0A182TS04 A0A0M9AA00 A0A1S4FKM6 Q16XH8 A0A154PPG4 A0A182GMZ9 A0A336KXF6 A0A1W4X3T6 U4UMC9 A0A0R3NKH1 Q293J4 A0A336M9T6 A0A2S2QK24 N6U0M0 A0A0L7QYM1 E9IZF0 A0A1A9WGW2 A0A310S6L6 A0A1I8PWQ2 A0A195CXR3 A0A151JQJ9 A0A151JXC9 A0A1B0G9S0 A0A3B0JSI1 A0A1A9UNH8 A0A1A9XQ49 A0A1A9ZDG9 A0A0R1E127 A0A0B4KHJ4 A0A0R1E5P0 Q9VDG0 A0A1I8M6W5 A0A0R1E1G1 B3P2Z8 A0A0R1E125 B4IKM6 A0A1B0BD95 A0A195AUI3 A0A1I8M6W0 B4PQ99 B4R1N9 A0A0P8Y0J0 A0A1W4VZS9 A0A1W4VLP2 B3M2X7 B4M4V0 A0A158P169 A0A0L0BTW6 A0A0J7P079 B4JSY1 E2BE63 A0A151WVZ6 B4KE24 E2AYZ0
A0A2J7PBF2 A0A2J7PBE9 A0A2J7RTB7 A0A2J7RTB8 A0A034UZR8 A0A2J7RTA3 A0A2J7RTA2 A0A0A9Y300 A0A2J7RTA7 A0A2J7RT97 A0A139WPL9 D6W7L1 A0A182SWG7 A0A1B0EYP6 E0VLA6 W8CAW5 A0A1J1IHV8 A0A2M4DP19 A0A232F8L3 A0A182V866 B0WUU8 A0A182IVR3 A0A1B6GIG4 A0A182YIP1 W5J456 A0A084VZI7 A0A182MNY4 A0A182LNA0 A0A2M4A0B6 A0A182FM39 A0A182NDL4 A0A1S4H4T4 A0A182W3T1 A0A182WR89 A0A182JRE6 A0A182PVS5 A0A182HX97 A0A182QB97 Q5TUD8 A0A182RBC6 A0A182TS04 A0A0M9AA00 A0A1S4FKM6 Q16XH8 A0A154PPG4 A0A182GMZ9 A0A336KXF6 A0A1W4X3T6 U4UMC9 A0A0R3NKH1 Q293J4 A0A336M9T6 A0A2S2QK24 N6U0M0 A0A0L7QYM1 E9IZF0 A0A1A9WGW2 A0A310S6L6 A0A1I8PWQ2 A0A195CXR3 A0A151JQJ9 A0A151JXC9 A0A1B0G9S0 A0A3B0JSI1 A0A1A9UNH8 A0A1A9XQ49 A0A1A9ZDG9 A0A0R1E127 A0A0B4KHJ4 A0A0R1E5P0 Q9VDG0 A0A1I8M6W5 A0A0R1E1G1 B3P2Z8 A0A0R1E125 B4IKM6 A0A1B0BD95 A0A195AUI3 A0A1I8M6W0 B4PQ99 B4R1N9 A0A0P8Y0J0 A0A1W4VZS9 A0A1W4VLP2 B3M2X7 B4M4V0 A0A158P169 A0A0L0BTW6 A0A0J7P079 B4JSY1 E2BE63 A0A151WVZ6 B4KE24 E2AYZ0
PDB
3VKH
E-value=1.14434e-19,
Score=233
Ontologies
GO
PANTHER
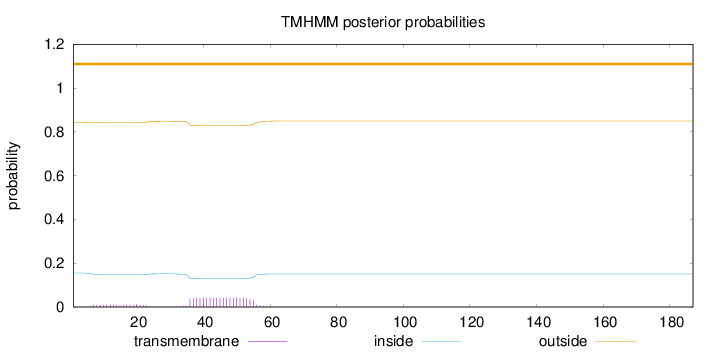
Topology
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.03234
Exp number, first 60 AAs:
1.02902
Total prob of N-in:
0.15572
outside
1 - 187
Population Genetic Test Statistics
Pi
82.308757
Theta
187.141398
Tajima's D
-1.781948
CLR
1443.525233
CSRT
0.0303984800759962
Interpretation
Possibly Positive selection
