Gene
KWMTBOMO02979
Pre Gene Modal
BGIBMGA003625
Annotation
PREDICTED:_uncharacterized_protein_LOC101737074_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.377
Sequence
CDS
ATGGTGTCTAGACGTCGAATATTATCGCGGTCACGCGACAACCTGTCGCAAACACTCACTCAGGAAGAGGAGGAAGACGTATGGTATCAAAAGGATAAGCTTTATAAGGAACATATACAGGAAGTGCTGGACAAATGGACACAAATCGACGACGAGATATGGGCGAAGGTGATTGTTTTCGAGAAGAATCGTCGCGTTGCCAAAGCGTATGCGCGAGCGCCGGTGCTCACGATTAATGGGTCCGATGACGGCTTTGACGGCATGAGAATTGGTCTATGCGGCTTCGATAATCCACATAGAGATCAGAAGACGGATGAATTGAAAAGACACGTAGGACAGGGTGTCAAAATCAAAATGGACGACGCCGGTAATATACTCATCAGACGCTACTCCAAGAGTAGCGTGTTTGTGAAGAGTACTGCAGCCTCTAGCAATGAAGAGACGGCCATCGGACAAGATATACTCAAACTGCCCGGATATTCGTTGGAGCAGGAAAAGATTTTTAAGTTGTTCGACATGAAGAAGTTCCAGTCAAACGTGAACCGGGAGTTGCGGCGCGCGTATCCGGACCGAAGAAGGCTTGAGACCCAGTGTCTCAGTGCTGTAGCATTTGTCAAGTCTGACAACGAACTCCTCGAGTGCCCCATCTGGGTGCTGGTCATCAACGTCGTCGCTATGGACATGCTGAAATCCAAGCTCCCCCCAATTCAAAGACCAATGGACATCAAGAACCGGCCCCGTATTCCAATTCCCGACGAGGATCCGTACAGCATCGCCAGTTCGAATGTCAACGGCTCCGGGTCAAGCGGCAGTTCAGGAGGCTACGGACTCGGCAACGGTCACAACGGACACGGAGTCGTCGCGGCTACTAGGGAGCAGCTGATGATGCAGATGGGTCAAAGGCGAGGAGAGAGACCACCTAATCTACCGCCCAGGGAGAATGGATATGGACCAGCTGATATTCCGAAGCCGGATTATGATGACATCGAGGCTAACGGTCGCGGTCCAGCGCAGTTTCCGAGAGGAAAATCAGACAAAGGCAAAGAAAAGAAATATGACGACCCCTACTATTGCGGGCTACGCGCCCGTGTGCCAAACTTCGTGAAGGCGAACGGAGCTAAAGTCCTCCCGACGATGACGGAGCGGCTCACGCTGAAGGAGGTGCCGGTGCCGAGCAAGCGGTTCAGCGTGGCGCACCCCCACGGCGCCCCTTACCCCCCGCACCCGCACATGCCCCCCGCTGCTCTCTGGCACGCGCGCTCCTACGAAAGCGGAATCGACGGAGATCATTACGATCCATACGCGTTGTACGGGCGGCTTCCGGTGCCTGGTCGTGGCTTCCCTCCGCCTCCCGCGCCTCATGCACGACACATGTTCATTGGAGATTGGGATTAA
Protein
MVSRRRILSRSRDNLSQTLTQEEEEDVWYQKDKLYKEHIQEVLDKWTQIDDEIWAKVIVFEKNRRVAKAYARAPVLTINGSDDGFDGMRIGLCGFDNPHRDQKTDELKRHVGQGVKIKMDDAGNILIRRYSKSSVFVKSTAASSNEETAIGQDILKLPGYSLEQEKIFKLFDMKKFQSNVNRELRRAYPDRRRLETQCLSAVAFVKSDNELLECPIWVLVINVVAMDMLKSKLPPIQRPMDIKNRPRIPIPDEDPYSIASSNVNGSGSSGSSGGYGLGNGHNGHGVVAATREQLMMQMGQRRGERPPNLPPRENGYGPADIPKPDYDDIEANGRGPAQFPRGKSDKGKEKKYDDPYYCGLRARVPNFVKANGAKVLPTMTERLTLKEVPVPSKRFSVAHPHGAPYPPHPHMPPAALWHARSYESGIDGDHYDPYALYGRLPVPGRGFPPPPAPHARHMFIGDWD
Summary
Uniprot
A0A212FGG9
Q7QJC4
B4HNV3
A0A1L8DMF8
B4MYM7
A0A0J9RAJ9
+ More
A1Z8R1 B4MDX2 B5E186 B4H591 B4J9V9 B4KSD6 Q7YU38 A1Z8R0 B4QC53 A0A0J9TZ81 A0A0M4E7Z2 B4P5Z0 A0A3B0J1U7 B3NSA8 B3MG41 A0A1W4VDS2 A0A0K8VFT6 A0A0Q5VPH7 A0A0R1DQQ7 A0A0P9AEL7 A0A182J2W2 A0A1W4VCW1 A0A1Y1L5S8 Q17LA6 A0A182XYS8 A0A182MRC8 A0A182QGH5 D6W7H1 A0A1J1IK81 A0A1Y1L9P0 A0A182UDY8 A0A182V2E2 A0A336JW18 A0A182KR80 A0A0L7LCZ0 A0A0A9XBX9 A0A2M4BNN5 A0A0K8SU61 A0A023F5F0 E2B0I0 A0A195CYG1 E9JAT5 A0A195EIU7 A0A2M4BNW1 A0A195FC59 A0A2S2QU85 A0A1B6MTQ3 F4W8G2 A0A026X300 A0A0L7RH77 E2BW45 A0A2S2QWC5 A0A2H8TSB9 A0A2S2PQQ7 A0A154P5S1 J9JNH1 A0A1B6KLA3 K7JBD2 A0A1B6FFC8 A0A151X714 A0A1B6G4T1 A0A232FK11 A0A0M9A4Z3 A0A146MC99 A0A2A3E1V8 V9IDW1 A0A310SHB8 A0A195BA16 A0A087ZQ44 E0VUT4 A0A0C9R5H1 A0A0P5SIJ1 A0A0P5H874 A0A0P5B3S6 A0A0P5PZQ6 A0A0P6FYK6 A0A0P5RA30 A0A0P5M5J7 A0A0P6ET66 A0A0P5TJM7 A0A0P5NYV8 A0A0P5M2I6 A0A162PGW1
A1Z8R1 B4MDX2 B5E186 B4H591 B4J9V9 B4KSD6 Q7YU38 A1Z8R0 B4QC53 A0A0J9TZ81 A0A0M4E7Z2 B4P5Z0 A0A3B0J1U7 B3NSA8 B3MG41 A0A1W4VDS2 A0A0K8VFT6 A0A0Q5VPH7 A0A0R1DQQ7 A0A0P9AEL7 A0A182J2W2 A0A1W4VCW1 A0A1Y1L5S8 Q17LA6 A0A182XYS8 A0A182MRC8 A0A182QGH5 D6W7H1 A0A1J1IK81 A0A1Y1L9P0 A0A182UDY8 A0A182V2E2 A0A336JW18 A0A182KR80 A0A0L7LCZ0 A0A0A9XBX9 A0A2M4BNN5 A0A0K8SU61 A0A023F5F0 E2B0I0 A0A195CYG1 E9JAT5 A0A195EIU7 A0A2M4BNW1 A0A195FC59 A0A2S2QU85 A0A1B6MTQ3 F4W8G2 A0A026X300 A0A0L7RH77 E2BW45 A0A2S2QWC5 A0A2H8TSB9 A0A2S2PQQ7 A0A154P5S1 J9JNH1 A0A1B6KLA3 K7JBD2 A0A1B6FFC8 A0A151X714 A0A1B6G4T1 A0A232FK11 A0A0M9A4Z3 A0A146MC99 A0A2A3E1V8 V9IDW1 A0A310SHB8 A0A195BA16 A0A087ZQ44 E0VUT4 A0A0C9R5H1 A0A0P5SIJ1 A0A0P5H874 A0A0P5B3S6 A0A0P5PZQ6 A0A0P6FYK6 A0A0P5RA30 A0A0P5M5J7 A0A0P6ET66 A0A0P5TJM7 A0A0P5NYV8 A0A0P5M2I6 A0A162PGW1
Pubmed
22118469
12364791
14747013
17210077
17994087
22936249
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18057021 15632085 26109357 26109356 17550304 28004739 17510324 25244985 18362917 19820115 20966253 26227816 25401762 25474469 20798317 21282665 21719571 24508170 30249741 20075255 28648823 26823975 20566863
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18057021 15632085 26109357 26109356 17550304 28004739 17510324 25244985 18362917 19820115 20966253 26227816 25401762 25474469 20798317 21282665 21719571 24508170 30249741 20075255 28648823 26823975 20566863
EMBL
AGBW02008676
OWR52820.1
AAAB01008807
EAA04134.4
CH480816
EDW47468.1
+ More
GFDF01006438 JAV07646.1 CH963894 EDW77216.1 CM002911 KMY93061.1 AE013599 AAF58603.2 CH940662 EDW58737.1 KRF78442.1 CM000071 EDY69474.1 CH479210 EDW32927.1 CH916367 EDW01523.1 CH933808 EDW08418.1 BT010005 AAQ22474.1 BT050452 AAM68700.2 ACJ13159.1 CM000362 EDX06684.1 KMY93060.1 CP012524 ALC40734.1 CM000158 EDW90865.1 OUUW01000001 SPP74987.1 CH954179 EDV56410.1 CH902619 EDV36736.1 GDHF01014530 GDHF01002056 JAI37784.1 JAI50258.1 KQS62867.1 KQS62868.1 KRJ99500.1 KRJ99501.1 KPU76390.1 KPU76391.1 GEZM01063668 JAV69029.1 CH477217 EAT47469.1 AXCM01001831 AXCN02000384 KQ971307 EFA11288.1 CVRI01000051 CRK99486.1 GEZM01063667 JAV69030.1 UFQS01000004 UFQT01000004 SSW96834.1 SSX17221.1 JTDY01001619 KOB73337.1 GBHO01025378 GBHO01025377 GBHO01025374 GBHO01025373 GBRD01009084 GBRD01009083 JAG18226.1 JAG18227.1 JAG18230.1 JAG18231.1 JAG56737.1 GGFJ01005549 MBW54690.1 GBRD01009085 GBRD01009082 JAG56739.1 GBBI01002349 JAC16363.1 GL444608 EFN60822.1 KQ977110 KYN05693.1 GL770679 EFZ10047.1 KQ978881 KYN27822.1 GGFJ01005327 MBW54468.1 KQ981693 KYN37634.1 GGMS01011887 MBY81090.1 GEBQ01000679 JAT39298.1 GL887908 EGI69453.1 KK107021 QOIP01000011 EZA62406.1 RLU16614.1 KQ414592 KOC70215.1 GL451091 EFN80077.1 GGMS01012760 MBY81963.1 GFXV01004956 MBW16761.1 GGMR01019182 MBY31801.1 KQ434809 KZC06480.1 ABLF02029248 ABLF02029249 GEBQ01027761 JAT12216.1 AAZX01003235 GECZ01020871 GECZ01014264 JAS48898.1 JAS55505.1 KQ982472 KYQ56088.1 GECZ01012325 GECZ01009037 JAS57444.1 JAS60732.1 NNAY01000097 OXU31002.1 KQ435756 KOX75959.1 GDHC01001255 JAQ17374.1 KZ288473 PBC25312.1 JR040740 JR040741 AEY59269.1 AEY59270.1 KQ761155 OAD58164.1 KQ976537 KYM81378.1 DS235793 EEB17140.1 GBYB01003185 JAG72952.1 GDIP01240334 GDIQ01123336 GDIQ01095286 JAI83067.1 JAL56440.1 GDIQ01231496 JAK20229.1 GDIP01194688 JAJ28714.1 GDIQ01123334 JAL28392.1 GDIP01240335 GDIQ01051775 JAI83066.1 JAN42962.1 GDIQ01123335 JAL28391.1 GDIQ01163270 JAK88455.1 GDIQ01065809 JAN28928.1 GDIP01125811 JAL77903.1 GDIQ01142786 JAL08940.1 GDIQ01161967 JAK89758.1 LRGB01000512 KZS18840.1
GFDF01006438 JAV07646.1 CH963894 EDW77216.1 CM002911 KMY93061.1 AE013599 AAF58603.2 CH940662 EDW58737.1 KRF78442.1 CM000071 EDY69474.1 CH479210 EDW32927.1 CH916367 EDW01523.1 CH933808 EDW08418.1 BT010005 AAQ22474.1 BT050452 AAM68700.2 ACJ13159.1 CM000362 EDX06684.1 KMY93060.1 CP012524 ALC40734.1 CM000158 EDW90865.1 OUUW01000001 SPP74987.1 CH954179 EDV56410.1 CH902619 EDV36736.1 GDHF01014530 GDHF01002056 JAI37784.1 JAI50258.1 KQS62867.1 KQS62868.1 KRJ99500.1 KRJ99501.1 KPU76390.1 KPU76391.1 GEZM01063668 JAV69029.1 CH477217 EAT47469.1 AXCM01001831 AXCN02000384 KQ971307 EFA11288.1 CVRI01000051 CRK99486.1 GEZM01063667 JAV69030.1 UFQS01000004 UFQT01000004 SSW96834.1 SSX17221.1 JTDY01001619 KOB73337.1 GBHO01025378 GBHO01025377 GBHO01025374 GBHO01025373 GBRD01009084 GBRD01009083 JAG18226.1 JAG18227.1 JAG18230.1 JAG18231.1 JAG56737.1 GGFJ01005549 MBW54690.1 GBRD01009085 GBRD01009082 JAG56739.1 GBBI01002349 JAC16363.1 GL444608 EFN60822.1 KQ977110 KYN05693.1 GL770679 EFZ10047.1 KQ978881 KYN27822.1 GGFJ01005327 MBW54468.1 KQ981693 KYN37634.1 GGMS01011887 MBY81090.1 GEBQ01000679 JAT39298.1 GL887908 EGI69453.1 KK107021 QOIP01000011 EZA62406.1 RLU16614.1 KQ414592 KOC70215.1 GL451091 EFN80077.1 GGMS01012760 MBY81963.1 GFXV01004956 MBW16761.1 GGMR01019182 MBY31801.1 KQ434809 KZC06480.1 ABLF02029248 ABLF02029249 GEBQ01027761 JAT12216.1 AAZX01003235 GECZ01020871 GECZ01014264 JAS48898.1 JAS55505.1 KQ982472 KYQ56088.1 GECZ01012325 GECZ01009037 JAS57444.1 JAS60732.1 NNAY01000097 OXU31002.1 KQ435756 KOX75959.1 GDHC01001255 JAQ17374.1 KZ288473 PBC25312.1 JR040740 JR040741 AEY59269.1 AEY59270.1 KQ761155 OAD58164.1 KQ976537 KYM81378.1 DS235793 EEB17140.1 GBYB01003185 JAG72952.1 GDIP01240334 GDIQ01123336 GDIQ01095286 JAI83067.1 JAL56440.1 GDIQ01231496 JAK20229.1 GDIP01194688 JAJ28714.1 GDIQ01123334 JAL28392.1 GDIP01240335 GDIQ01051775 JAI83066.1 JAN42962.1 GDIQ01123335 JAL28391.1 GDIQ01163270 JAK88455.1 GDIQ01065809 JAN28928.1 GDIP01125811 JAL77903.1 GDIQ01142786 JAL08940.1 GDIQ01161967 JAK89758.1 LRGB01000512 KZS18840.1
Proteomes
UP000007151
UP000007062
UP000001292
UP000007798
UP000000803
UP000008792
+ More
UP000001819 UP000008744 UP000001070 UP000009192 UP000000304 UP000092553 UP000002282 UP000268350 UP000008711 UP000007801 UP000192221 UP000075880 UP000008820 UP000076408 UP000075883 UP000075886 UP000007266 UP000183832 UP000075902 UP000075882 UP000037510 UP000000311 UP000078542 UP000078492 UP000078541 UP000007755 UP000053097 UP000279307 UP000053825 UP000008237 UP000076502 UP000007819 UP000002358 UP000075809 UP000215335 UP000053105 UP000242457 UP000078540 UP000005203 UP000009046 UP000076858
UP000001819 UP000008744 UP000001070 UP000009192 UP000000304 UP000092553 UP000002282 UP000268350 UP000008711 UP000007801 UP000192221 UP000075880 UP000008820 UP000076408 UP000075883 UP000075886 UP000007266 UP000183832 UP000075902 UP000075882 UP000037510 UP000000311 UP000078542 UP000078492 UP000078541 UP000007755 UP000053097 UP000279307 UP000053825 UP000008237 UP000076502 UP000007819 UP000002358 UP000075809 UP000215335 UP000053105 UP000242457 UP000078540 UP000005203 UP000009046 UP000076858
Pfam
PF03166 MH2
Interpro
SUPFAM
SSF49879
SSF49879
Gene 3D
ProteinModelPortal
A0A212FGG9
Q7QJC4
B4HNV3
A0A1L8DMF8
B4MYM7
A0A0J9RAJ9
+ More
A1Z8R1 B4MDX2 B5E186 B4H591 B4J9V9 B4KSD6 Q7YU38 A1Z8R0 B4QC53 A0A0J9TZ81 A0A0M4E7Z2 B4P5Z0 A0A3B0J1U7 B3NSA8 B3MG41 A0A1W4VDS2 A0A0K8VFT6 A0A0Q5VPH7 A0A0R1DQQ7 A0A0P9AEL7 A0A182J2W2 A0A1W4VCW1 A0A1Y1L5S8 Q17LA6 A0A182XYS8 A0A182MRC8 A0A182QGH5 D6W7H1 A0A1J1IK81 A0A1Y1L9P0 A0A182UDY8 A0A182V2E2 A0A336JW18 A0A182KR80 A0A0L7LCZ0 A0A0A9XBX9 A0A2M4BNN5 A0A0K8SU61 A0A023F5F0 E2B0I0 A0A195CYG1 E9JAT5 A0A195EIU7 A0A2M4BNW1 A0A195FC59 A0A2S2QU85 A0A1B6MTQ3 F4W8G2 A0A026X300 A0A0L7RH77 E2BW45 A0A2S2QWC5 A0A2H8TSB9 A0A2S2PQQ7 A0A154P5S1 J9JNH1 A0A1B6KLA3 K7JBD2 A0A1B6FFC8 A0A151X714 A0A1B6G4T1 A0A232FK11 A0A0M9A4Z3 A0A146MC99 A0A2A3E1V8 V9IDW1 A0A310SHB8 A0A195BA16 A0A087ZQ44 E0VUT4 A0A0C9R5H1 A0A0P5SIJ1 A0A0P5H874 A0A0P5B3S6 A0A0P5PZQ6 A0A0P6FYK6 A0A0P5RA30 A0A0P5M5J7 A0A0P6ET66 A0A0P5TJM7 A0A0P5NYV8 A0A0P5M2I6 A0A162PGW1
A1Z8R1 B4MDX2 B5E186 B4H591 B4J9V9 B4KSD6 Q7YU38 A1Z8R0 B4QC53 A0A0J9TZ81 A0A0M4E7Z2 B4P5Z0 A0A3B0J1U7 B3NSA8 B3MG41 A0A1W4VDS2 A0A0K8VFT6 A0A0Q5VPH7 A0A0R1DQQ7 A0A0P9AEL7 A0A182J2W2 A0A1W4VCW1 A0A1Y1L5S8 Q17LA6 A0A182XYS8 A0A182MRC8 A0A182QGH5 D6W7H1 A0A1J1IK81 A0A1Y1L9P0 A0A182UDY8 A0A182V2E2 A0A336JW18 A0A182KR80 A0A0L7LCZ0 A0A0A9XBX9 A0A2M4BNN5 A0A0K8SU61 A0A023F5F0 E2B0I0 A0A195CYG1 E9JAT5 A0A195EIU7 A0A2M4BNW1 A0A195FC59 A0A2S2QU85 A0A1B6MTQ3 F4W8G2 A0A026X300 A0A0L7RH77 E2BW45 A0A2S2QWC5 A0A2H8TSB9 A0A2S2PQQ7 A0A154P5S1 J9JNH1 A0A1B6KLA3 K7JBD2 A0A1B6FFC8 A0A151X714 A0A1B6G4T1 A0A232FK11 A0A0M9A4Z3 A0A146MC99 A0A2A3E1V8 V9IDW1 A0A310SHB8 A0A195BA16 A0A087ZQ44 E0VUT4 A0A0C9R5H1 A0A0P5SIJ1 A0A0P5H874 A0A0P5B3S6 A0A0P5PZQ6 A0A0P6FYK6 A0A0P5RA30 A0A0P5M5J7 A0A0P6ET66 A0A0P5TJM7 A0A0P5NYV8 A0A0P5M2I6 A0A162PGW1
PDB
4R9P
E-value=6.10457e-115,
Score=1060
Ontologies
GO
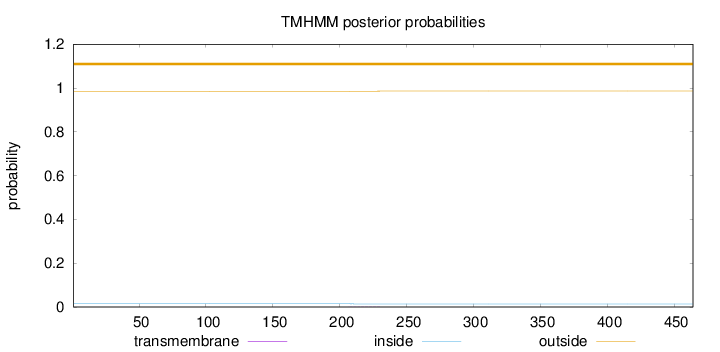
Topology
Length:
464
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02276
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01469
outside
1 - 464
Population Genetic Test Statistics
Pi
230.897451
Theta
181.643913
Tajima's D
0.982138
CLR
0.47404
CSRT
0.660516974151292
Interpretation
Uncertain
