Gene
KWMTBOMO02978 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003579
Annotation
putative_serine_protease_K12H4.7_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 1.798
Sequence
CDS
ATGTTGTTTATTGGGACTAACATAATTCTCGCTTGTATTAGTGTTGGTTTTGGATTTGTACTGAACGGACTCGGAAATAGGTACAGACATTTGCTTCAAGATATAAATTCAACGCTGACGTCATTTGATACATCGTCTATCAAAATCAAATGGATAGAGCAGCCTTTAGATCATTTCGATAAAAAAAATATAAATACTTGGAATATGAGGTATATGGAAAGATTAGATAAATGGAAGAAAGATGGTCCTATATACCTATTTATGTACGGTGAGGGTGAAGCAAGTCAGGTATTTCTTCAAACTGGTATTTTGTATGAATTAGCCGAGGAAACAAACGGGGCTATGTTGCTATCTGAACACAGATACTATGGAAAGAGCAAGCCTTTTGAAGAACTCACAACTGAAAATCTAAAGTTCCTAAGTTCAAGGCAAGCACTCGCAGATACTGCTAGATTGTTGGAATTAGTCAAATCAGAACCTCAATATGAAAATTCGAAAATTGTTGTTGTTGGAGGATCATACGCAGGTAATTTGGCTGCTTGGATGAGACTACTGTATCCCAATTTAATTAATGCTGCTATAGCCAGTAGCGCTCCGGTATTAGCAAAGAAAGATTTCTACGAATATTTAGAACACGTCAGCGATGATTTTGAACAACACGGTACTGAAGACTGTATGGAGAAAATTAATGAAATTTTTACTACATACGAGAAACGATTTCGAACAGTAGATGGCATTAAACAACTTAAAAAGGAGGAGTACATCTGTGAAGACACCGATATGACGAAAGTCGAAAATCAGCAATTATTCTTTTTGGATAAAAGCTCTGCATTTATGTACATTGCACAATACGGGAACCCAGATGTCATCAAGACGTATTGCGAAGATATTAAAAATACCTCTCTTGTATTTTCTTCTAAAGTAGATGAAAACGATTTTTACGACACTAAAAATAATTGTTACGATTATGAATTTGAAGAAATGATTCAAAACACGAAAGAAATAGACTGGCTCGTTTCATGGACATATCAAACTTGTACTGAATTCAGTTACTTTCCGAGCACATCATCAAACAAACATCCTTTTACTGATAATATAAGTATGGACCTGTTTTATAAAATGTGTACTAAAACTTTTGGTCCGAACTTTGACGAAAAGAGAATAGAGGCTGGGGTTATGGAAACCAATATGATGTATGGTGGATTAACTCCTAATGTAACAAAGGTGGTATTTGTTAACGGTGATCTAGACCCATATCATAGATTAAGTATTCTAGAAGATTTATCTTATGAAGCTCCAGCAAAAGTTATTCCACGTACATCCCATTGCAGAGATTTATTTTCAAACAGATCTGGCGACCCTGAAGAACTTATCGAAGCCAGGTATTATATAAAATATTTGATAAAAAAATGGATAGGTCTTGGTGACTATAACAAACCAAATGAAATAAAGCACACAACCTAG
Protein
MLFIGTNIILACISVGFGFVLNGLGNRYRHLLQDINSTLTSFDTSSIKIKWIEQPLDHFDKKNINTWNMRYMERLDKWKKDGPIYLFMYGEGEASQVFLQTGILYELAEETNGAMLLSEHRYYGKSKPFEELTTENLKFLSSRQALADTARLLELVKSEPQYENSKIVVVGGSYAGNLAAWMRLLYPNLINAAIASSAPVLAKKDFYEYLEHVSDDFEQHGTEDCMEKINEIFTTYEKRFRTVDGIKQLKKEEYICEDTDMTKVENQQLFFLDKSSAFMYIAQYGNPDVIKTYCEDIKNTSLVFSSKVDENDFYDTKNNCYDYEFEEMIQNTKEIDWLVSWTYQTCTEFSYFPSTSSNKHPFTDNISMDLFYKMCTKTFGPNFDEKRIEAGVMETNMMYGGLTPNVTKVVFVNGDLDPYHRLSILEDLSYEAPAKVIPRTSHCRDLFSNRSGDPEELIEARYYIKYLIKKWIGLGDYNKPNEIKHTT
Summary
Uniprot
H9J242
A0A2A4IXM3
A0A0L7LD48
A0A2A4IXD9
A0A2H1W1T1
A0A0N0PDT7
+ More
A0A194PQR5 A0A220K8N5 A0A212FGE5 A0A154P900 A0A026WSC7 A0A0M9A6W0 F4WG05 A0A158NL17 E2BVA8 A0A0L7QS79 A0A195EFD4 A0A088A3L2 A0A151XBU0 A0A195F208 A0A195BHC6 K7IN08 A0A232EUP8 A0A195BXZ0 E9IN24 A0A1Q3FIL9 A0A182SU00 A0A1Q3FIJ9 T1DU14 A0A182RA60 A0A182K7G7 W5J436 A0A084W051 A0A1Q3FI84 A0A182PRT2 A0A0N7ZA73 A0A182FS07 A0A182YQ28 A0A182NQQ2 A0A182X9F2 A0A182QSB3 Q7PX68 Q16MC1 A0A182JJ09 A0A182TDU4 E2ASK6 A0A182L8L9 B0XF46 A0A182HKD8 A0A2T7NCZ5 A0A1Q3FJA3 A0A182V4Z4 A0A151XFV2 A0A2A3EUH5 B0XF47 A0A023EV78 A0A182VVY5 A0A1B6JIG6 A0A151JZ82 F4WUJ0 A0A1B6IAX2 A0A195CTE8 A0A1B6I0B9 A0A195E9L6 A0A2J7QIH2 A0A2G8JM64 E9J3M5 A0A1L8DP91 E9FVW6 A0A336N030 A0A1B6CAA8 A0A158NFD3 A0A2C9K4W9 A0A210PQI8 E0VXG1 A0A087ZYG3 B4KD56 A0A0B7A1H0 A0A1V1FNB5 A0A0V0VSF4 V3ZZH9 A0A2B4R661 A0A0T6AW67 A0A034WBZ8 A0A3P8V5J4 X1WJF0 A0A1J1I7I0 A0A0V0S9A4 A0A1B6JRB9 A0A1Y3EBE0 B4KD57 E2C1R7 A0A0P5KXN6 A0A1L8DPC5 A0A1L8DPN7 A0A1B6H0P6 A0A0P5ACM5 A0A0V1AFK6 A0A182MCQ3 A0A067RFD1
A0A194PQR5 A0A220K8N5 A0A212FGE5 A0A154P900 A0A026WSC7 A0A0M9A6W0 F4WG05 A0A158NL17 E2BVA8 A0A0L7QS79 A0A195EFD4 A0A088A3L2 A0A151XBU0 A0A195F208 A0A195BHC6 K7IN08 A0A232EUP8 A0A195BXZ0 E9IN24 A0A1Q3FIL9 A0A182SU00 A0A1Q3FIJ9 T1DU14 A0A182RA60 A0A182K7G7 W5J436 A0A084W051 A0A1Q3FI84 A0A182PRT2 A0A0N7ZA73 A0A182FS07 A0A182YQ28 A0A182NQQ2 A0A182X9F2 A0A182QSB3 Q7PX68 Q16MC1 A0A182JJ09 A0A182TDU4 E2ASK6 A0A182L8L9 B0XF46 A0A182HKD8 A0A2T7NCZ5 A0A1Q3FJA3 A0A182V4Z4 A0A151XFV2 A0A2A3EUH5 B0XF47 A0A023EV78 A0A182VVY5 A0A1B6JIG6 A0A151JZ82 F4WUJ0 A0A1B6IAX2 A0A195CTE8 A0A1B6I0B9 A0A195E9L6 A0A2J7QIH2 A0A2G8JM64 E9J3M5 A0A1L8DP91 E9FVW6 A0A336N030 A0A1B6CAA8 A0A158NFD3 A0A2C9K4W9 A0A210PQI8 E0VXG1 A0A087ZYG3 B4KD56 A0A0B7A1H0 A0A1V1FNB5 A0A0V0VSF4 V3ZZH9 A0A2B4R661 A0A0T6AW67 A0A034WBZ8 A0A3P8V5J4 X1WJF0 A0A1J1I7I0 A0A0V0S9A4 A0A1B6JRB9 A0A1Y3EBE0 B4KD57 E2C1R7 A0A0P5KXN6 A0A1L8DPC5 A0A1L8DPN7 A0A1B6H0P6 A0A0P5ACM5 A0A0V1AFK6 A0A182MCQ3 A0A067RFD1
Pubmed
19121390
26227816
26354079
28630352
22118469
24508170
+ More
21719571 21347285 20798317 20075255 28648823 21282665 20920257 23761445 24438588 25244985 12364791 14747013 17210077 17510324 20966253 24945155 29023486 21292972 15562597 28812685 20566863 17994087 28410430 23254933 25348373 24487278 24845553
21719571 21347285 20798317 20075255 28648823 21282665 20920257 23761445 24438588 25244985 12364791 14747013 17210077 17510324 20966253 24945155 29023486 21292972 15562597 28812685 20566863 17994087 28410430 23254933 25348373 24487278 24845553
EMBL
BABH01007474
NWSH01005531
PCG64148.1
JTDY01001619
KOB73334.1
PCG64146.1
+ More
ODYU01005773 SOQ46987.1 KQ460226 KPJ16489.1 KQ459597 KPI95084.1 MF319736 ASJ26450.1 AGBW02008676 OWR52821.1 KQ434846 KZC08379.1 KK107111 EZA58932.1 KQ435724 KOX78447.1 GL888128 EGI66772.1 ADTU01019141 GL450824 EFN80400.1 KQ414758 KOC61505.1 KQ978957 KYN26958.1 KQ982320 KYQ57780.1 KQ981866 KYN34129.1 KQ976488 KYM83571.1 NNAY01002105 OXU22074.1 KQ978501 KYM93462.1 GL764272 EFZ18031.1 GFDL01007637 JAV27408.1 GFDL01007656 JAV27389.1 GAMD01000569 JAB01022.1 ADMH02002101 ETN59152.1 ATLV01019089 KE525262 KFB43595.1 GFDL01007775 JAV27270.1 GDRN01103633 JAI58048.1 AXCN02001508 AAAB01008987 EAA01781.5 CH477867 EAT35480.1 GL442330 EFN63566.1 DS232895 EDS26459.1 APCN01000826 PZQS01000014 PVD19043.1 GFDL01007441 JAV27604.1 KQ982182 KYQ59185.1 KZ288187 PBC34711.1 EDS26460.1 GAPW01001214 GEHC01000588 JAC12384.1 JAV47057.1 GECU01008716 JAS98990.1 KQ981410 KYN41918.1 GL888365 EGI62155.1 GECU01023610 JAS84096.1 KQ977279 KYN03978.1 GECU01027362 JAS80344.1 KQ979440 KYN21502.1 NEVH01013592 PNF28388.1 MRZV01001613 PIK36813.1 GL768066 EFZ12594.1 GFDF01005781 JAV08303.1 GL732525 EFX88622.1 UFQS01001724 UFQT01001724 SSX12010.1 SSX31528.1 GEDC01026876 JAS10422.1 ADTU01014274 NEDP02005555 OWF38769.1 DS235832 EEB18067.1 CH933806 EDW14838.2 HACG01027807 HACG01027808 CEK74672.1 CEK74673.1 FX985297 BAX07310.1 JYDN01000011 KRX66431.1 KB203083 ESO86391.1 LSMT01000886 PFX13834.1 LJIG01022644 KRT79472.1 GAKP01007297 GAKP01007296 JAC51655.1 ABLF02021908 ABLF02021912 CVRI01000042 CRK95698.1 JYDL01000026 KRX23231.1 GECU01005922 JAT01785.1 LVZM01017505 OUC42462.1 EDW14839.2 GL451975 EFN78126.1 GDIQ01186157 GDIP01089752 JAK65568.1 JAM13963.1 GFDF01005785 JAV08299.1 GFDF01005784 JAV08300.1 GECZ01001513 JAS68256.1 GDIP01200934 JAJ22468.1 JYDQ01000003 KRY23476.1 AXCM01003685 KK852542 KDR21723.1
ODYU01005773 SOQ46987.1 KQ460226 KPJ16489.1 KQ459597 KPI95084.1 MF319736 ASJ26450.1 AGBW02008676 OWR52821.1 KQ434846 KZC08379.1 KK107111 EZA58932.1 KQ435724 KOX78447.1 GL888128 EGI66772.1 ADTU01019141 GL450824 EFN80400.1 KQ414758 KOC61505.1 KQ978957 KYN26958.1 KQ982320 KYQ57780.1 KQ981866 KYN34129.1 KQ976488 KYM83571.1 NNAY01002105 OXU22074.1 KQ978501 KYM93462.1 GL764272 EFZ18031.1 GFDL01007637 JAV27408.1 GFDL01007656 JAV27389.1 GAMD01000569 JAB01022.1 ADMH02002101 ETN59152.1 ATLV01019089 KE525262 KFB43595.1 GFDL01007775 JAV27270.1 GDRN01103633 JAI58048.1 AXCN02001508 AAAB01008987 EAA01781.5 CH477867 EAT35480.1 GL442330 EFN63566.1 DS232895 EDS26459.1 APCN01000826 PZQS01000014 PVD19043.1 GFDL01007441 JAV27604.1 KQ982182 KYQ59185.1 KZ288187 PBC34711.1 EDS26460.1 GAPW01001214 GEHC01000588 JAC12384.1 JAV47057.1 GECU01008716 JAS98990.1 KQ981410 KYN41918.1 GL888365 EGI62155.1 GECU01023610 JAS84096.1 KQ977279 KYN03978.1 GECU01027362 JAS80344.1 KQ979440 KYN21502.1 NEVH01013592 PNF28388.1 MRZV01001613 PIK36813.1 GL768066 EFZ12594.1 GFDF01005781 JAV08303.1 GL732525 EFX88622.1 UFQS01001724 UFQT01001724 SSX12010.1 SSX31528.1 GEDC01026876 JAS10422.1 ADTU01014274 NEDP02005555 OWF38769.1 DS235832 EEB18067.1 CH933806 EDW14838.2 HACG01027807 HACG01027808 CEK74672.1 CEK74673.1 FX985297 BAX07310.1 JYDN01000011 KRX66431.1 KB203083 ESO86391.1 LSMT01000886 PFX13834.1 LJIG01022644 KRT79472.1 GAKP01007297 GAKP01007296 JAC51655.1 ABLF02021908 ABLF02021912 CVRI01000042 CRK95698.1 JYDL01000026 KRX23231.1 GECU01005922 JAT01785.1 LVZM01017505 OUC42462.1 EDW14839.2 GL451975 EFN78126.1 GDIQ01186157 GDIP01089752 JAK65568.1 JAM13963.1 GFDF01005785 JAV08299.1 GFDF01005784 JAV08300.1 GECZ01001513 JAS68256.1 GDIP01200934 JAJ22468.1 JYDQ01000003 KRY23476.1 AXCM01003685 KK852542 KDR21723.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000076502 UP000053097 UP000053105 UP000007755 UP000005205 UP000008237 UP000053825 UP000078492 UP000005203 UP000075809 UP000078541 UP000078540 UP000002358 UP000215335 UP000078542 UP000075901 UP000075900 UP000075881 UP000000673 UP000030765 UP000075885 UP000069272 UP000076408 UP000075884 UP000076407 UP000075886 UP000007062 UP000008820 UP000075880 UP000075902 UP000000311 UP000075882 UP000002320 UP000075840 UP000245119 UP000075903 UP000242457 UP000075920 UP000235965 UP000230750 UP000000305 UP000076420 UP000242188 UP000009046 UP000009192 UP000054681 UP000030746 UP000225706 UP000265120 UP000007819 UP000183832 UP000054630 UP000243006 UP000054783 UP000075883 UP000027135
UP000076502 UP000053097 UP000053105 UP000007755 UP000005205 UP000008237 UP000053825 UP000078492 UP000005203 UP000075809 UP000078541 UP000078540 UP000002358 UP000215335 UP000078542 UP000075901 UP000075900 UP000075881 UP000000673 UP000030765 UP000075885 UP000069272 UP000076408 UP000075884 UP000076407 UP000075886 UP000007062 UP000008820 UP000075880 UP000075902 UP000000311 UP000075882 UP000002320 UP000075840 UP000245119 UP000075903 UP000242457 UP000075920 UP000235965 UP000230750 UP000000305 UP000076420 UP000242188 UP000009046 UP000009192 UP000054681 UP000030746 UP000225706 UP000265120 UP000007819 UP000183832 UP000054630 UP000243006 UP000054783 UP000075883 UP000027135
PRIDE
Pfam
PF05577 Peptidase_S28
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9J242
A0A2A4IXM3
A0A0L7LD48
A0A2A4IXD9
A0A2H1W1T1
A0A0N0PDT7
+ More
A0A194PQR5 A0A220K8N5 A0A212FGE5 A0A154P900 A0A026WSC7 A0A0M9A6W0 F4WG05 A0A158NL17 E2BVA8 A0A0L7QS79 A0A195EFD4 A0A088A3L2 A0A151XBU0 A0A195F208 A0A195BHC6 K7IN08 A0A232EUP8 A0A195BXZ0 E9IN24 A0A1Q3FIL9 A0A182SU00 A0A1Q3FIJ9 T1DU14 A0A182RA60 A0A182K7G7 W5J436 A0A084W051 A0A1Q3FI84 A0A182PRT2 A0A0N7ZA73 A0A182FS07 A0A182YQ28 A0A182NQQ2 A0A182X9F2 A0A182QSB3 Q7PX68 Q16MC1 A0A182JJ09 A0A182TDU4 E2ASK6 A0A182L8L9 B0XF46 A0A182HKD8 A0A2T7NCZ5 A0A1Q3FJA3 A0A182V4Z4 A0A151XFV2 A0A2A3EUH5 B0XF47 A0A023EV78 A0A182VVY5 A0A1B6JIG6 A0A151JZ82 F4WUJ0 A0A1B6IAX2 A0A195CTE8 A0A1B6I0B9 A0A195E9L6 A0A2J7QIH2 A0A2G8JM64 E9J3M5 A0A1L8DP91 E9FVW6 A0A336N030 A0A1B6CAA8 A0A158NFD3 A0A2C9K4W9 A0A210PQI8 E0VXG1 A0A087ZYG3 B4KD56 A0A0B7A1H0 A0A1V1FNB5 A0A0V0VSF4 V3ZZH9 A0A2B4R661 A0A0T6AW67 A0A034WBZ8 A0A3P8V5J4 X1WJF0 A0A1J1I7I0 A0A0V0S9A4 A0A1B6JRB9 A0A1Y3EBE0 B4KD57 E2C1R7 A0A0P5KXN6 A0A1L8DPC5 A0A1L8DPN7 A0A1B6H0P6 A0A0P5ACM5 A0A0V1AFK6 A0A182MCQ3 A0A067RFD1
A0A194PQR5 A0A220K8N5 A0A212FGE5 A0A154P900 A0A026WSC7 A0A0M9A6W0 F4WG05 A0A158NL17 E2BVA8 A0A0L7QS79 A0A195EFD4 A0A088A3L2 A0A151XBU0 A0A195F208 A0A195BHC6 K7IN08 A0A232EUP8 A0A195BXZ0 E9IN24 A0A1Q3FIL9 A0A182SU00 A0A1Q3FIJ9 T1DU14 A0A182RA60 A0A182K7G7 W5J436 A0A084W051 A0A1Q3FI84 A0A182PRT2 A0A0N7ZA73 A0A182FS07 A0A182YQ28 A0A182NQQ2 A0A182X9F2 A0A182QSB3 Q7PX68 Q16MC1 A0A182JJ09 A0A182TDU4 E2ASK6 A0A182L8L9 B0XF46 A0A182HKD8 A0A2T7NCZ5 A0A1Q3FJA3 A0A182V4Z4 A0A151XFV2 A0A2A3EUH5 B0XF47 A0A023EV78 A0A182VVY5 A0A1B6JIG6 A0A151JZ82 F4WUJ0 A0A1B6IAX2 A0A195CTE8 A0A1B6I0B9 A0A195E9L6 A0A2J7QIH2 A0A2G8JM64 E9J3M5 A0A1L8DP91 E9FVW6 A0A336N030 A0A1B6CAA8 A0A158NFD3 A0A2C9K4W9 A0A210PQI8 E0VXG1 A0A087ZYG3 B4KD56 A0A0B7A1H0 A0A1V1FNB5 A0A0V0VSF4 V3ZZH9 A0A2B4R661 A0A0T6AW67 A0A034WBZ8 A0A3P8V5J4 X1WJF0 A0A1J1I7I0 A0A0V0S9A4 A0A1B6JRB9 A0A1Y3EBE0 B4KD57 E2C1R7 A0A0P5KXN6 A0A1L8DPC5 A0A1L8DPN7 A0A1B6H0P6 A0A0P5ACM5 A0A0V1AFK6 A0A182MCQ3 A0A067RFD1
PDB
3N2Z
E-value=1.37315e-40,
Score=419
Ontologies
GO
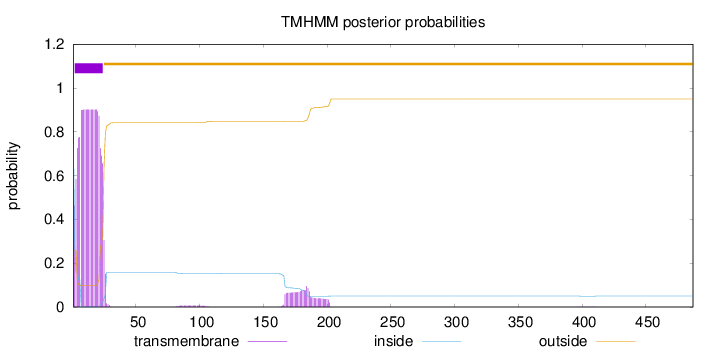
Topology
Length:
487
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.56349
Exp number, first 60 AAs:
19.373
Total prob of N-in:
0.74335
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 24
outside
25 - 487
Population Genetic Test Statistics
Pi
199.061508
Theta
176.072067
Tajima's D
0.515635
CLR
0.227714
CSRT
0.515974201289936
Interpretation
Uncertain
