Pre Gene Modal
BGIBMGA003627
Annotation
hypothetical_protein_SINV_14695?_partial_[Solenopsis_invicta]
Location in the cell
Cytoplasmic Reliability : 1.243 Mitochondrial Reliability : 1.386
Sequence
CDS
ATGGGGAAGCTTCTCTCGAAGATATTCGGTAACAAGGAGATGAGAATACTGATGCTGGGACTCGACGCTGCTGGAAAGACTACCATTTTATATAAGTTAAAATTAGGTCAGTCTGTCACGACAATACCGACAGTGGGCTTCAACGTTGAAACCGTAACGTATAAGAACGTCAAATTTAATGTCTGGGACGTCGGGGGGCAGGACAAAATACGGCCGCTGTGGAGGCATTACTACACAGGCACTCAGGGCCTGATATTCGTTGTGGACTGCGCGGACCGCGACCGTATCGACGAGGCCCGCCAGGAATTGCACAGGATCATAAACGACAGGGAGATGCGGGACGCCATCATACTCATATTCGCCAACAAACAGGATCTACCTGACGCGATGAAACCGCACGAGATACAAGAGAAACTGGGGCTGACCCGCATCCGGGACCGGAACTGGTACGTGCAGCCGTCGTGCGCGACGACCGGCGACGGACTGTACGAGGGGCTCACGTGGCTCACCAGCAATCATAAGTTATGA
Protein
MGKLLSKIFGNKEMRILMLGLDAAGKTTILYKLKLGQSVTTIPTVGFNVETVTYKNVKFNVWDVGGQDKIRPLWRHYYTGTQGLIFVVDCADRDRIDEARQELHRIINDREMRDAIILIFANKQDLPDAMKPHEIQEKLGLTRIRDRNWYVQPSCATTGDGLYEGLTWLTSNHKL
Summary
Similarity
Belongs to the small GTPase superfamily. Arf family.
Uniprot
A0A1B6IBG6
E9J4R7
A0A232F5N9
A0A151IE20
A0A154PRU9
A0A0L7QUF4
+ More
F4X4R6 T1E337 A0A151HYB4 A0A2A3ELN0 A0A2J7RKN0 A0A0M9A7W3 A0A1B6GVC7 A0A195ET55 A0A158NSF1 E2AX37 A0A165VG46 A0A1B6BZJ5 V9IBQ5 A0A1B6MLP4 A0A0C9RFG9 A0A026WZ49 A0A1L8DVQ9 A0A2A4JBB6 A0A0N1PJH4 A0A1E1WH37 A0A2H1V215 A0A1B0DPE8 A0A194PNS6 T1IZ00 E2C8J1 Q1HQT1 A0A1Q3G1Z1 A0A182Y826 A0A182M9U3 A0A182PIP0 A0A182R0S5 A0A182V4T4 A0A182N9F8 A0A182K1D3 A0A182WBD3 A0A182TGF9 B0X466 E0VHZ9 A0A182L3U1 A0A182WWD0 A0A182RJF5 A0A023EI61 A0A182I6K9 A0A067QUD3 A0A2M3ZAC1 A0A2M4AQ24 A0A087ZM30 A0A084W680 A0A182IKT2 A0A182FGR5 U5ELG0 A0A1S3CYR1 Q17BN9 S4PEW9 A0A212FKH6 A0A1S3HU42 V5GG98 D6W7V9 A0A0T6B3G5 A0A1Y1LHF4 A0A1W4WJG0 T1IF41 A0A023F5V5 A0A147A565 A0A1A8GBV6 A0A1A7WZ49 A0A1A8I852 A0A1A8DE18 A0A1A8B203 A0A0K8TQE6 A0A087YQL1 I3KYB4 A0A060XDW9 B5X494 M4AYI0 A0A3B4EZP9 A0A3P9C0R3 A0A2I4BLY9 W5NMF1 C1BM37 A0A060WSN7 A0A315VV46 A0A2U9CIX8 A0A1A8SCS7 A0A1A8NBH0 A0A1A8K0R8 A0A0S7L8Z1 A0A1A7X2M0 Q6P2U5 A0A1W4ZLZ6 A0A2D0RJS5 A0A1A8ECM3 A0A1A8HLZ5 A0A1S3MB69
F4X4R6 T1E337 A0A151HYB4 A0A2A3ELN0 A0A2J7RKN0 A0A0M9A7W3 A0A1B6GVC7 A0A195ET55 A0A158NSF1 E2AX37 A0A165VG46 A0A1B6BZJ5 V9IBQ5 A0A1B6MLP4 A0A0C9RFG9 A0A026WZ49 A0A1L8DVQ9 A0A2A4JBB6 A0A0N1PJH4 A0A1E1WH37 A0A2H1V215 A0A1B0DPE8 A0A194PNS6 T1IZ00 E2C8J1 Q1HQT1 A0A1Q3G1Z1 A0A182Y826 A0A182M9U3 A0A182PIP0 A0A182R0S5 A0A182V4T4 A0A182N9F8 A0A182K1D3 A0A182WBD3 A0A182TGF9 B0X466 E0VHZ9 A0A182L3U1 A0A182WWD0 A0A182RJF5 A0A023EI61 A0A182I6K9 A0A067QUD3 A0A2M3ZAC1 A0A2M4AQ24 A0A087ZM30 A0A084W680 A0A182IKT2 A0A182FGR5 U5ELG0 A0A1S3CYR1 Q17BN9 S4PEW9 A0A212FKH6 A0A1S3HU42 V5GG98 D6W7V9 A0A0T6B3G5 A0A1Y1LHF4 A0A1W4WJG0 T1IF41 A0A023F5V5 A0A147A565 A0A1A8GBV6 A0A1A7WZ49 A0A1A8I852 A0A1A8DE18 A0A1A8B203 A0A0K8TQE6 A0A087YQL1 I3KYB4 A0A060XDW9 B5X494 M4AYI0 A0A3B4EZP9 A0A3P9C0R3 A0A2I4BLY9 W5NMF1 C1BM37 A0A060WSN7 A0A315VV46 A0A2U9CIX8 A0A1A8SCS7 A0A1A8NBH0 A0A1A8K0R8 A0A0S7L8Z1 A0A1A7X2M0 Q6P2U5 A0A1W4ZLZ6 A0A2D0RJS5 A0A1A8ECM3 A0A1A8HLZ5 A0A1S3MB69
Pubmed
EMBL
GECU01027933
GECU01023421
GECU01018902
GECU01018590
GECU01013330
JAS79773.1
+ More
JAS84285.1 JAS88804.1 JAS89116.1 JAS94376.1 GL768112 EFZ12210.1 NNAY01000872 OXU26124.1 KQ977894 KYM98948.1 KQ435078 KZC14467.1 KQ414735 KOC62273.1 GL888679 EGI58489.1 GALA01000725 JAA94127.1 KQ976730 KYM76465.1 KZ288215 PBC32610.1 NEVH01002717 PNF41394.1 KQ435716 KOX79055.1 GECZ01003412 JAS66357.1 KQ981993 KYN31064.1 ADTU01024892 GL443520 EFN61987.1 KT984807 AMZ00359.1 GEDC01030913 JAS06385.1 JR037960 JR037961 AEY58057.1 AEY58058.1 GEBQ01031883 GEBQ01026198 GEBQ01010826 GEBQ01003164 JAT08094.1 JAT13779.1 JAT29151.1 JAT36813.1 GBYB01011862 GBYB01011863 JAG81629.1 JAG81630.1 KK107061 QOIP01000014 EZA61335.1 RLU15117.1 GFDF01003560 JAV10524.1 NWSH01002144 PCG69056.1 KQ460226 KPJ16486.1 GDQN01004776 JAT86278.1 ODYU01000300 SOQ34806.1 AJVK01008050 AJVK01008051 AJVK01008052 KQ459597 KPI95081.1 JH431701 GL453666 EFN75721.1 DQ440363 ABF18396.1 GFDL01001269 JAV33776.1 AXCM01016960 AXCN02000560 DS232330 EDS40143.1 DS235174 EEB13005.1 JXUM01040422 JXUM01091369 JXUM01091370 JXUM01091371 JXUM01091372 JXUM01091373 GAPW01005003 KQ563907 KQ561244 JAC08595.1 KXJ73086.1 KXJ79236.1 APCN01003657 KK853253 KDR09315.1 GGFM01004735 MBW25486.1 GGFK01009556 MBW42877.1 GGFL01004227 MBW68405.1 ATLV01020755 KE525307 KFB45724.1 AXCP01001420 GANO01001421 JAB58450.1 CH477319 EAT43674.1 GAIX01004307 JAA88253.1 AGBW02008033 OWR54241.1 GALX01005482 JAB62984.1 KQ971307 EFA11143.1 LJIG01015994 KRT81932.1 GEZM01055251 GEZM01055250 JAV73064.1 ACPB03011946 GBBI01002148 JAC16564.1 GCES01012629 JAR73694.1 HAEC01000476 SBQ68553.1 HADW01009575 SBP10975.1 HAED01007050 HAEE01011226 SBQ93230.1 HADZ01014723 HAEA01003168 SBQ31648.1 HADY01022261 HAEJ01001873 SBP60746.1 GDAI01001503 JAI16100.1 AYCK01015200 AERX01030270 FR905227 FR908043 CDQ77412.1 CDQ87628.1 BT045863 ACI34125.1 AHAT01003726 BT075666 ACO10090.1 FR904590 CDQ67605.1 NHOQ01001000 PWA27466.1 CP026259 AWP16545.1 HAEH01016664 HAEI01013269 SBS15738.1 HAEF01005384 HAEG01001912 SBR66204.1 HAED01017250 HAEE01005792 SBR25812.1 GBYX01180866 GBYX01180864 JAO85921.1 HADW01010876 HADX01015309 SBP12276.1 CT027758 CU693483 BC064293 BC154500 AAH64293.1 AAI54501.1 HADZ01007437 HAEA01014876 SBQ43356.1 HAEB01002134 HAEC01015992 SBQ84213.1
JAS84285.1 JAS88804.1 JAS89116.1 JAS94376.1 GL768112 EFZ12210.1 NNAY01000872 OXU26124.1 KQ977894 KYM98948.1 KQ435078 KZC14467.1 KQ414735 KOC62273.1 GL888679 EGI58489.1 GALA01000725 JAA94127.1 KQ976730 KYM76465.1 KZ288215 PBC32610.1 NEVH01002717 PNF41394.1 KQ435716 KOX79055.1 GECZ01003412 JAS66357.1 KQ981993 KYN31064.1 ADTU01024892 GL443520 EFN61987.1 KT984807 AMZ00359.1 GEDC01030913 JAS06385.1 JR037960 JR037961 AEY58057.1 AEY58058.1 GEBQ01031883 GEBQ01026198 GEBQ01010826 GEBQ01003164 JAT08094.1 JAT13779.1 JAT29151.1 JAT36813.1 GBYB01011862 GBYB01011863 JAG81629.1 JAG81630.1 KK107061 QOIP01000014 EZA61335.1 RLU15117.1 GFDF01003560 JAV10524.1 NWSH01002144 PCG69056.1 KQ460226 KPJ16486.1 GDQN01004776 JAT86278.1 ODYU01000300 SOQ34806.1 AJVK01008050 AJVK01008051 AJVK01008052 KQ459597 KPI95081.1 JH431701 GL453666 EFN75721.1 DQ440363 ABF18396.1 GFDL01001269 JAV33776.1 AXCM01016960 AXCN02000560 DS232330 EDS40143.1 DS235174 EEB13005.1 JXUM01040422 JXUM01091369 JXUM01091370 JXUM01091371 JXUM01091372 JXUM01091373 GAPW01005003 KQ563907 KQ561244 JAC08595.1 KXJ73086.1 KXJ79236.1 APCN01003657 KK853253 KDR09315.1 GGFM01004735 MBW25486.1 GGFK01009556 MBW42877.1 GGFL01004227 MBW68405.1 ATLV01020755 KE525307 KFB45724.1 AXCP01001420 GANO01001421 JAB58450.1 CH477319 EAT43674.1 GAIX01004307 JAA88253.1 AGBW02008033 OWR54241.1 GALX01005482 JAB62984.1 KQ971307 EFA11143.1 LJIG01015994 KRT81932.1 GEZM01055251 GEZM01055250 JAV73064.1 ACPB03011946 GBBI01002148 JAC16564.1 GCES01012629 JAR73694.1 HAEC01000476 SBQ68553.1 HADW01009575 SBP10975.1 HAED01007050 HAEE01011226 SBQ93230.1 HADZ01014723 HAEA01003168 SBQ31648.1 HADY01022261 HAEJ01001873 SBP60746.1 GDAI01001503 JAI16100.1 AYCK01015200 AERX01030270 FR905227 FR908043 CDQ77412.1 CDQ87628.1 BT045863 ACI34125.1 AHAT01003726 BT075666 ACO10090.1 FR904590 CDQ67605.1 NHOQ01001000 PWA27466.1 CP026259 AWP16545.1 HAEH01016664 HAEI01013269 SBS15738.1 HAEF01005384 HAEG01001912 SBR66204.1 HAED01017250 HAEE01005792 SBR25812.1 GBYX01180866 GBYX01180864 JAO85921.1 HADW01010876 HADX01015309 SBP12276.1 CT027758 CU693483 BC064293 BC154500 AAH64293.1 AAI54501.1 HADZ01007437 HAEA01014876 SBQ43356.1 HAEB01002134 HAEC01015992 SBQ84213.1
Proteomes
UP000215335
UP000078542
UP000076502
UP000053825
UP000007755
UP000078540
+ More
UP000242457 UP000235965 UP000053105 UP000078541 UP000005205 UP000000311 UP000053097 UP000279307 UP000218220 UP000053240 UP000092462 UP000053268 UP000008237 UP000076408 UP000075883 UP000075885 UP000075886 UP000075903 UP000075884 UP000075881 UP000075920 UP000075902 UP000002320 UP000009046 UP000075882 UP000076407 UP000075900 UP000069940 UP000249989 UP000075840 UP000027135 UP000030765 UP000075880 UP000069272 UP000079169 UP000008820 UP000007151 UP000085678 UP000007266 UP000192223 UP000015103 UP000265000 UP000028760 UP000005207 UP000193380 UP000087266 UP000002852 UP000261460 UP000265160 UP000192220 UP000018468 UP000246464 UP000000437 UP000192224 UP000221080
UP000242457 UP000235965 UP000053105 UP000078541 UP000005205 UP000000311 UP000053097 UP000279307 UP000218220 UP000053240 UP000092462 UP000053268 UP000008237 UP000076408 UP000075883 UP000075885 UP000075886 UP000075903 UP000075884 UP000075881 UP000075920 UP000075902 UP000002320 UP000009046 UP000075882 UP000076407 UP000075900 UP000069940 UP000249989 UP000075840 UP000027135 UP000030765 UP000075880 UP000069272 UP000079169 UP000008820 UP000007151 UP000085678 UP000007266 UP000192223 UP000015103 UP000265000 UP000028760 UP000005207 UP000193380 UP000087266 UP000002852 UP000261460 UP000265160 UP000192220 UP000018468 UP000246464 UP000000437 UP000192224 UP000221080
Pfam
PF00025 Arf
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A1B6IBG6
E9J4R7
A0A232F5N9
A0A151IE20
A0A154PRU9
A0A0L7QUF4
+ More
F4X4R6 T1E337 A0A151HYB4 A0A2A3ELN0 A0A2J7RKN0 A0A0M9A7W3 A0A1B6GVC7 A0A195ET55 A0A158NSF1 E2AX37 A0A165VG46 A0A1B6BZJ5 V9IBQ5 A0A1B6MLP4 A0A0C9RFG9 A0A026WZ49 A0A1L8DVQ9 A0A2A4JBB6 A0A0N1PJH4 A0A1E1WH37 A0A2H1V215 A0A1B0DPE8 A0A194PNS6 T1IZ00 E2C8J1 Q1HQT1 A0A1Q3G1Z1 A0A182Y826 A0A182M9U3 A0A182PIP0 A0A182R0S5 A0A182V4T4 A0A182N9F8 A0A182K1D3 A0A182WBD3 A0A182TGF9 B0X466 E0VHZ9 A0A182L3U1 A0A182WWD0 A0A182RJF5 A0A023EI61 A0A182I6K9 A0A067QUD3 A0A2M3ZAC1 A0A2M4AQ24 A0A087ZM30 A0A084W680 A0A182IKT2 A0A182FGR5 U5ELG0 A0A1S3CYR1 Q17BN9 S4PEW9 A0A212FKH6 A0A1S3HU42 V5GG98 D6W7V9 A0A0T6B3G5 A0A1Y1LHF4 A0A1W4WJG0 T1IF41 A0A023F5V5 A0A147A565 A0A1A8GBV6 A0A1A7WZ49 A0A1A8I852 A0A1A8DE18 A0A1A8B203 A0A0K8TQE6 A0A087YQL1 I3KYB4 A0A060XDW9 B5X494 M4AYI0 A0A3B4EZP9 A0A3P9C0R3 A0A2I4BLY9 W5NMF1 C1BM37 A0A060WSN7 A0A315VV46 A0A2U9CIX8 A0A1A8SCS7 A0A1A8NBH0 A0A1A8K0R8 A0A0S7L8Z1 A0A1A7X2M0 Q6P2U5 A0A1W4ZLZ6 A0A2D0RJS5 A0A1A8ECM3 A0A1A8HLZ5 A0A1S3MB69
F4X4R6 T1E337 A0A151HYB4 A0A2A3ELN0 A0A2J7RKN0 A0A0M9A7W3 A0A1B6GVC7 A0A195ET55 A0A158NSF1 E2AX37 A0A165VG46 A0A1B6BZJ5 V9IBQ5 A0A1B6MLP4 A0A0C9RFG9 A0A026WZ49 A0A1L8DVQ9 A0A2A4JBB6 A0A0N1PJH4 A0A1E1WH37 A0A2H1V215 A0A1B0DPE8 A0A194PNS6 T1IZ00 E2C8J1 Q1HQT1 A0A1Q3G1Z1 A0A182Y826 A0A182M9U3 A0A182PIP0 A0A182R0S5 A0A182V4T4 A0A182N9F8 A0A182K1D3 A0A182WBD3 A0A182TGF9 B0X466 E0VHZ9 A0A182L3U1 A0A182WWD0 A0A182RJF5 A0A023EI61 A0A182I6K9 A0A067QUD3 A0A2M3ZAC1 A0A2M4AQ24 A0A087ZM30 A0A084W680 A0A182IKT2 A0A182FGR5 U5ELG0 A0A1S3CYR1 Q17BN9 S4PEW9 A0A212FKH6 A0A1S3HU42 V5GG98 D6W7V9 A0A0T6B3G5 A0A1Y1LHF4 A0A1W4WJG0 T1IF41 A0A023F5V5 A0A147A565 A0A1A8GBV6 A0A1A7WZ49 A0A1A8I852 A0A1A8DE18 A0A1A8B203 A0A0K8TQE6 A0A087YQL1 I3KYB4 A0A060XDW9 B5X494 M4AYI0 A0A3B4EZP9 A0A3P9C0R3 A0A2I4BLY9 W5NMF1 C1BM37 A0A060WSN7 A0A315VV46 A0A2U9CIX8 A0A1A8SCS7 A0A1A8NBH0 A0A1A8K0R8 A0A0S7L8Z1 A0A1A7X2M0 Q6P2U5 A0A1W4ZLZ6 A0A2D0RJS5 A0A1A8ECM3 A0A1A8HLZ5 A0A1S3MB69
PDB
6BBQ
E-value=1.94238e-99,
Score=921
Ontologies
GO
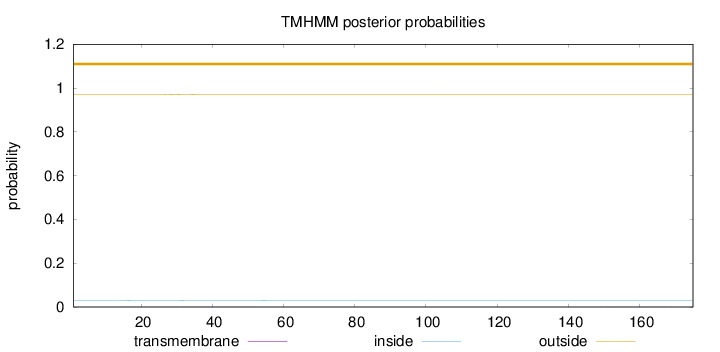
Topology
Length:
175
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02591
Exp number, first 60 AAs:
0.0201
Total prob of N-in:
0.02945
outside
1 - 175
Population Genetic Test Statistics
Pi
214.187825
Theta
173.987902
Tajima's D
0.392016
CLR
0.885844
CSRT
0.485475726213689
Interpretation
Uncertain
