Gene
KWMTBOMO02965
Annotation
PREDICTED:_uncharacterized_protein_LOC101738091_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.848
Sequence
CDS
ATGGATAATGGAACAAAAGAGGAAAAGAATCGTATTTGGGATTACGCTGTTTGTTTCGGGACAGTTATCACATTCATAGCCGGCATTGGTCACGTAAATTCATTTGGATTGATTTACAGAGATTTTATGATCCACACTCAATCCACTACTAAGTCGTTGACAACTGCCAATGGCGTATTTGCCATAATGCTAGCAATCGGAGGTATTATTCTCAATATTATTACCAAGAAATTAACGCTAAGACAGTGCGGTCTGATCGGTTCTTGTATATTTTCGACCGGATCGTTTCTAACAATACTGATCTCGAATACTAATCATCTACCGTTCACGTTTGGTGTGCTGCAAGGTATCGGCTTTGGGATCCTGGTCCCGGTGTGCTATTCAACATTGAATTTACTTTTCACGACAAAAAGAACAACAGTTATGAGCATAATAAAATCATTACAAGGAATAATATTAATGTGGTATCCGCAAGTTATTAAACAGACGTTATCTGTTTACGGCTTCAGAGGGACGCTGCTAATCATTTTTGGAATTTCATTGCATACTTTTCCCGGAATGTTATTAATGAAGACAGAAAATAAGGATACTGGAAAGAGACTAAGAACAGCAAAAAATATAGAGATCGAAACAGACAAACAAATAAAATTAGAGTTATTAAATAACGATGAACAGAAACAACAAATTCTAAGCGATAATGGAACTGAATACAAGACGTCGAAGAAAAATAGGTTTCACTTCTTAGAAGTACTGAATCTGAGATGCCTTAAAGATCCCGTCTATTGCAATATTTGTGTCGGTCAGAGTTTTATGAATTTCTCCGATTTGACTTTCTTCATTTTACAGCCGATGCTTTTGTTTCAATACGGATACGACAAGGCGCAAGTCGCTACTTGTATTACAATTGGGACAGGAGCTGACGTGGCGGGCCGACTGGCCCTGGCCATGATTAGTGGTGCGACACATATTAACACCAGATTATTGTACTACGTTGCTACGCTGCTCACGTTGGTCGTAAGATTAGTTTTACTTCAAATCCATGAATTCATCTGGGTAGCAGCCATTACAGCTGTGCTGGGTATACTCCGAGCATGGATCCACATAGCCAGTCCTCTGGTAATTTCTAATCACGTCACCCACGAAGACTTTGCTGGGGCATACGCTCTGTTCATGCTTGCTGCGGGCGTGGTCAATTTAACGTGTTCACCTTTTATAGGCATGATTAAGGATTCGTATGAAGACTATGTACCTGCATTCTATGCGTTGTCAGCTAGCTGTTTGCCTTGTCTGATAATGTGGCCTATCGAATTAGTGTCAAGACGAAAATGA
Protein
MDNGTKEEKNRIWDYAVCFGTVITFIAGIGHVNSFGLIYRDFMIHTQSTTKSLTTANGVFAIMLAIGGIILNIITKKLTLRQCGLIGSCIFSTGSFLTILISNTNHLPFTFGVLQGIGFGILVPVCYSTLNLLFTTKRTTVMSIIKSLQGIILMWYPQVIKQTLSVYGFRGTLLIIFGISLHTFPGMLLMKTENKDTGKRLRTAKNIEIETDKQIKLELLNNDEQKQQILSDNGTEYKTSKKNRFHFLEVLNLRCLKDPVYCNICVGQSFMNFSDLTFFILQPMLLFQYGYDKAQVATCITIGTGADVAGRLALAMISGATHINTRLLYYVATLLTLVVRLVLLQIHEFIWVAAITAVLGILRAWIHIASPLVISNHVTHEDFAGAYALFMLAAGVVNLTCSPFIGMIKDSYEDYVPAFYALSASCLPCLIMWPIELVSRRK
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
A0A2A4JER7
A0A2H1VVT2
A0A194PP43
A0A0N1IHC4
A0A212F801
Q17DF9
+ More
A0A2W1BJL7 A0A182GDP8 W5JLX9 B0XIY5 A0A1Q3FJ07 A0A2H1VXL3 A0A182W0Y1 A0A212FNW6 A0A182RBY8 A0A182QJX4 A0A182F9C7 A0A084WTD4 A0A182MSX0 A0A182IPW6 U5ESW5 A0A182TYU4 Q7QCE2 A0A182VGB7 A0A182XAS2 A0A182HSL8 A0A182NZV0 A0A2A4JNU3 A0A2H1V1M4 A0A182Y6H4 A0A182NRI8 A0A182K474 A0A1A9ZB14 A8JUR0 A0A1A9UZI4 A0A1I8M485 A0A0Q5TAQ5 A0A1A9W694 B4I741 A0A194R4V2 A0A1I8Q6B1 A0A1L8DEC6 A0A1B0GJR3 A0A1B0DHB5 A0A1L8EFW6 Q9VWJ2 B3N140 D2A1Q7 H9IV66 A0A1L8DDT9 A0A1Y1N8V7 A0A067RH10 A0A212F810 T1E9I8 A0A0L7KRT6 A0A194PVC1 A0A2A4K764 A0A088ABH2 A0A1W4XGX8 B4NUQ6 A0A2W1BKS8 N6TRZ7 N6T394 A0A0N1IT74 A0A1W4WYB4 A0A2W1BIC5 A0A2A3E764 A0A151X7M3 A0A1W4WZD5 A0A195DLX4 A0A088ABH3 A0A1Y1MBK1 A0A1Y1MGP1
A0A2W1BJL7 A0A182GDP8 W5JLX9 B0XIY5 A0A1Q3FJ07 A0A2H1VXL3 A0A182W0Y1 A0A212FNW6 A0A182RBY8 A0A182QJX4 A0A182F9C7 A0A084WTD4 A0A182MSX0 A0A182IPW6 U5ESW5 A0A182TYU4 Q7QCE2 A0A182VGB7 A0A182XAS2 A0A182HSL8 A0A182NZV0 A0A2A4JNU3 A0A2H1V1M4 A0A182Y6H4 A0A182NRI8 A0A182K474 A0A1A9ZB14 A8JUR0 A0A1A9UZI4 A0A1I8M485 A0A0Q5TAQ5 A0A1A9W694 B4I741 A0A194R4V2 A0A1I8Q6B1 A0A1L8DEC6 A0A1B0GJR3 A0A1B0DHB5 A0A1L8EFW6 Q9VWJ2 B3N140 D2A1Q7 H9IV66 A0A1L8DDT9 A0A1Y1N8V7 A0A067RH10 A0A212F810 T1E9I8 A0A0L7KRT6 A0A194PVC1 A0A2A4K764 A0A088ABH2 A0A1W4XGX8 B4NUQ6 A0A2W1BKS8 N6TRZ7 N6T394 A0A0N1IT74 A0A1W4WYB4 A0A2W1BIC5 A0A2A3E764 A0A151X7M3 A0A1W4WZD5 A0A195DLX4 A0A088ABH3 A0A1Y1MBK1 A0A1Y1MGP1
Pubmed
EMBL
NWSH01001768
PCG70198.1
ODYU01004743
SOQ44945.1
KQ459597
KPI95077.1
+ More
KQ460226 KPJ16482.1 AGBW02009819 OWR49848.1 CH477295 EAT44425.1 KZ150104 PZC73457.1 JXUM01010240 KQ560302 KXJ83164.1 ADMH02001190 ETN63784.1 DS233409 EDS29793.1 GFDL01007537 JAV27508.1 SOQ44944.1 AGBW02004200 OWR55438.1 AXCN02001457 ATLV01026883 KE525420 KFB53478.1 AXCM01000202 GANO01003057 JAB56814.1 AAAB01008859 EAA08073.5 APCN01000295 NWSH01001018 PCG73092.1 ODYU01000236 SOQ34666.1 AE014298 ABW09455.2 CH954180 KQS30081.1 CH480823 EDW56139.1 KQ460761 KPJ12539.1 GFDF01009272 JAV04812.1 AJWK01022144 AJWK01022145 AJWK01022146 AJVK01060889 GFDG01001263 JAV17536.1 AAF48946.3 CH902650 EDV30075.2 KQ971338 EFA01508.1 BABH01000581 GFDF01009468 JAV04616.1 GEZM01012259 JAV93035.1 KK852511 KDR22318.1 OWR49849.1 GAMD01001307 JAB00284.1 JTDY01006442 KOB66012.1 KPI95075.1 NWSH01000106 PCG79492.1 CH984177 EDX16703.1 PZC73456.1 APGK01054138 APGK01054139 KB741247 ENN72030.1 ENN72028.1 KQ435878 KOX69908.1 PZC73455.1 KZ288347 PBC27540.1 KQ982446 KYQ56363.1 KQ980734 KYN13878.1 GEZM01036000 JAV83043.1 GEZM01035998 JAV83046.1
KQ460226 KPJ16482.1 AGBW02009819 OWR49848.1 CH477295 EAT44425.1 KZ150104 PZC73457.1 JXUM01010240 KQ560302 KXJ83164.1 ADMH02001190 ETN63784.1 DS233409 EDS29793.1 GFDL01007537 JAV27508.1 SOQ44944.1 AGBW02004200 OWR55438.1 AXCN02001457 ATLV01026883 KE525420 KFB53478.1 AXCM01000202 GANO01003057 JAB56814.1 AAAB01008859 EAA08073.5 APCN01000295 NWSH01001018 PCG73092.1 ODYU01000236 SOQ34666.1 AE014298 ABW09455.2 CH954180 KQS30081.1 CH480823 EDW56139.1 KQ460761 KPJ12539.1 GFDF01009272 JAV04812.1 AJWK01022144 AJWK01022145 AJWK01022146 AJVK01060889 GFDG01001263 JAV17536.1 AAF48946.3 CH902650 EDV30075.2 KQ971338 EFA01508.1 BABH01000581 GFDF01009468 JAV04616.1 GEZM01012259 JAV93035.1 KK852511 KDR22318.1 OWR49849.1 GAMD01001307 JAB00284.1 JTDY01006442 KOB66012.1 KPI95075.1 NWSH01000106 PCG79492.1 CH984177 EDX16703.1 PZC73456.1 APGK01054138 APGK01054139 KB741247 ENN72030.1 ENN72028.1 KQ435878 KOX69908.1 PZC73455.1 KZ288347 PBC27540.1 KQ982446 KYQ56363.1 KQ980734 KYN13878.1 GEZM01036000 JAV83043.1 GEZM01035998 JAV83046.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000008820
UP000069940
+ More
UP000249989 UP000000673 UP000002320 UP000075920 UP000075900 UP000075886 UP000069272 UP000030765 UP000075883 UP000075880 UP000075902 UP000007062 UP000075903 UP000076407 UP000075840 UP000075885 UP000076408 UP000075884 UP000075881 UP000092445 UP000000803 UP000078200 UP000095301 UP000008711 UP000091820 UP000001292 UP000095300 UP000092461 UP000092462 UP000007801 UP000007266 UP000005204 UP000027135 UP000037510 UP000005203 UP000192223 UP000000304 UP000019118 UP000053105 UP000242457 UP000075809 UP000078492
UP000249989 UP000000673 UP000002320 UP000075920 UP000075900 UP000075886 UP000069272 UP000030765 UP000075883 UP000075880 UP000075902 UP000007062 UP000075903 UP000076407 UP000075840 UP000075885 UP000076408 UP000075884 UP000075881 UP000092445 UP000000803 UP000078200 UP000095301 UP000008711 UP000091820 UP000001292 UP000095300 UP000092461 UP000092462 UP000007801 UP000007266 UP000005204 UP000027135 UP000037510 UP000005203 UP000192223 UP000000304 UP000019118 UP000053105 UP000242457 UP000075809 UP000078492
Pfam
Interpro
IPR020846
MFS_dom
+ More
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR011108 RMMBL
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR022712 Beta_Casp
IPR001279 Metallo-B-lactamas
IPR021718 CPSF73-100_C
IPR002403 Cyt_P450_E_grp-IV
IPR017972 Cyt_P450_CS
IPR001128 Cyt_P450
IPR036396 Cyt_P450_sf
IPR022127 STIMATE/YPL162C
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR016187 CTDL_fold
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR011108 RMMBL
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR022712 Beta_Casp
IPR001279 Metallo-B-lactamas
IPR021718 CPSF73-100_C
IPR002403 Cyt_P450_E_grp-IV
IPR017972 Cyt_P450_CS
IPR001128 Cyt_P450
IPR036396 Cyt_P450_sf
IPR022127 STIMATE/YPL162C
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR016187 CTDL_fold
Gene 3D
CDD
ProteinModelPortal
A0A2A4JER7
A0A2H1VVT2
A0A194PP43
A0A0N1IHC4
A0A212F801
Q17DF9
+ More
A0A2W1BJL7 A0A182GDP8 W5JLX9 B0XIY5 A0A1Q3FJ07 A0A2H1VXL3 A0A182W0Y1 A0A212FNW6 A0A182RBY8 A0A182QJX4 A0A182F9C7 A0A084WTD4 A0A182MSX0 A0A182IPW6 U5ESW5 A0A182TYU4 Q7QCE2 A0A182VGB7 A0A182XAS2 A0A182HSL8 A0A182NZV0 A0A2A4JNU3 A0A2H1V1M4 A0A182Y6H4 A0A182NRI8 A0A182K474 A0A1A9ZB14 A8JUR0 A0A1A9UZI4 A0A1I8M485 A0A0Q5TAQ5 A0A1A9W694 B4I741 A0A194R4V2 A0A1I8Q6B1 A0A1L8DEC6 A0A1B0GJR3 A0A1B0DHB5 A0A1L8EFW6 Q9VWJ2 B3N140 D2A1Q7 H9IV66 A0A1L8DDT9 A0A1Y1N8V7 A0A067RH10 A0A212F810 T1E9I8 A0A0L7KRT6 A0A194PVC1 A0A2A4K764 A0A088ABH2 A0A1W4XGX8 B4NUQ6 A0A2W1BKS8 N6TRZ7 N6T394 A0A0N1IT74 A0A1W4WYB4 A0A2W1BIC5 A0A2A3E764 A0A151X7M3 A0A1W4WZD5 A0A195DLX4 A0A088ABH3 A0A1Y1MBK1 A0A1Y1MGP1
A0A2W1BJL7 A0A182GDP8 W5JLX9 B0XIY5 A0A1Q3FJ07 A0A2H1VXL3 A0A182W0Y1 A0A212FNW6 A0A182RBY8 A0A182QJX4 A0A182F9C7 A0A084WTD4 A0A182MSX0 A0A182IPW6 U5ESW5 A0A182TYU4 Q7QCE2 A0A182VGB7 A0A182XAS2 A0A182HSL8 A0A182NZV0 A0A2A4JNU3 A0A2H1V1M4 A0A182Y6H4 A0A182NRI8 A0A182K474 A0A1A9ZB14 A8JUR0 A0A1A9UZI4 A0A1I8M485 A0A0Q5TAQ5 A0A1A9W694 B4I741 A0A194R4V2 A0A1I8Q6B1 A0A1L8DEC6 A0A1B0GJR3 A0A1B0DHB5 A0A1L8EFW6 Q9VWJ2 B3N140 D2A1Q7 H9IV66 A0A1L8DDT9 A0A1Y1N8V7 A0A067RH10 A0A212F810 T1E9I8 A0A0L7KRT6 A0A194PVC1 A0A2A4K764 A0A088ABH2 A0A1W4XGX8 B4NUQ6 A0A2W1BKS8 N6TRZ7 N6T394 A0A0N1IT74 A0A1W4WYB4 A0A2W1BIC5 A0A2A3E764 A0A151X7M3 A0A1W4WZD5 A0A195DLX4 A0A088ABH3 A0A1Y1MBK1 A0A1Y1MGP1
Ontologies
GO
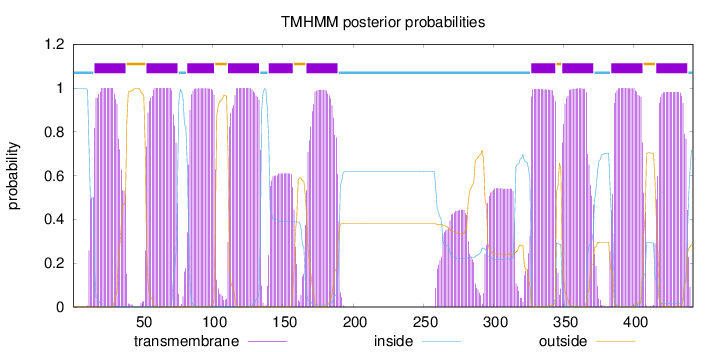
Topology
Length:
442
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
225.14985
Exp number, first 60 AAs:
29.97335
Total prob of N-in:
0.99724
POSSIBLE N-term signal
sequence
inside
1 - 15
TMhelix
16 - 38
outside
39 - 52
TMhelix
53 - 75
inside
76 - 81
TMhelix
82 - 101
outside
102 - 110
TMhelix
111 - 133
inside
134 - 139
TMhelix
140 - 157
outside
158 - 166
TMhelix
167 - 189
inside
190 - 326
TMhelix
327 - 344
outside
345 - 348
TMhelix
349 - 371
inside
372 - 383
TMhelix
384 - 406
outside
407 - 415
TMhelix
416 - 438
inside
439 - 442
Population Genetic Test Statistics
Pi
226.714074
Theta
177.535462
Tajima's D
0.624108
CLR
0.176124
CSRT
0.556172191390431
Interpretation
Uncertain
