Gene
KWMTBOMO02964
Pre Gene Modal
BGIBMGA003572
Annotation
PREDICTED:_monocarboxylate_transporter_9-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.939
Sequence
CDS
ATGAGTGTGGTTAAAGAAGAGTGTTGTGAAGAAGAGAAGTACGAATTAGTTCCTCCCGAAGGAGGCTGGGGCTACGTTGTCTGTGTGGGATTATCTGTAATTTTCATTGCTGGCACCGCGCATCAGCCTGTTTTTGGTTTTATATACAATGATTTTCTGGACGAACTTGGCGTCGGTACTGGCGCCGTTACAGTTGTATATGGAGTGTTTCAAGTCACATTAGCTATTGCCGGTTTTTCGGCGAACATAGCTTTGAAAAGTCTTTCATTAAGACAAGTCGGTCTTATAGGTGCCTTTATATACACACTAGCATCATTCCTAGCAATATTTGTAGTATCCACAACTCAGTTAATTGTCACAAACGGATTCCTACAGGGCCTCGGAATGGGTCTACTAATTCCGGTGTCGTATACTAGTTTTAACAGTTACTTTACAAAGAAAAAAGTGCTGTATTTGAGTCTGTGTAAGGCATCTATTGGATTGATAACGATGTTGTACCCGCTGTTTATAAAATTTACGATAACCCAATACGGGTTTAGAGGGACTCTAGCGATTATTTGTGCCATATCAGCTCACAGTATTTTCGGAGCTCTAGTCATGCATCCTGTACAATGGTATATGGTGAAGAAATTGAAAACGTGCGAAAAGATTGTCCTGATACCACCGACGCCGGCCATTGAGGCGGGAAAAGAAAAATTCCATTTCAATTCTAACACGACGGCATCCAAAGGAGCGGAAAACGCGAGATCAGTGGAAAACTTGTACGCAATCAAACGGACGGGGAGTCAAAAACCCACGGAATTGCTATTCGTTCCGAAATTAGCTGATTCCAGACGACTGAGTATACCGGTGGTTTTGATCCCAGGGGAAACGAAGGCTGATAGCAAATTTAATTCGAGCGATAATGTGTATTTAGAGAATTTGTATAAGGCAGCTTCGGTCTCCAGTTTGGGTAATTTCGGGAATATCGCAGCTGAACCAATGCCAGTTATCAAGAACAAGGAAGGAAAGTGGCAAACGATAATGGAATACTTGGATCTGACTTTACTCAAAGATTTGGTTTACGACAATATCATTGTCGGAATGGCTTTGACATTTTTCGCTGATCTAACTTTCTTCACTTTGGAGCCTTTATTCTTGGATAAAGTCAATTTGAATCGGAGCCAAATTGCTAACATAATAGCTTTAGGCGGTGCTACGGACATGTGCGCCAGATTGTTCCTGGGCGTGGCGGGACAGTTCTTCATTATGAATAGCCGGTATATGTTCTTCATAGGAGCCCTTTTTGCAGCCGCTTTTAGATTGGTTATGATACAATTCACAACATACCTACCTCTACTAATATTGACTGGTATACTCGGAGCCTTGAGGTCTTTGGTGCATATAGCGCAGCCTATAGTGATGGCTGAACACGTTCCCATAGAGCTGTACCCATCGGCTTATGGACTCTACATGTTACTCGCCGGAGCGATTAGTTTGAGTGTCGGACCCATGATAGGATTCATTCGGGACTTCACTGGTAGTTATGTGGTTGCTTTCGTTATGTTGGCTGTGTGTAATCTATGCTGCGTTGTTCCGTGGGCGATAGAGTTCATTTTTCGTTACAAAAAAGAAAGGAAACTCAGAAATACGGACGTGAGCTGA
Protein
MSVVKEECCEEEKYELVPPEGGWGYVVCVGLSVIFIAGTAHQPVFGFIYNDFLDELGVGTGAVTVVYGVFQVTLAIAGFSANIALKSLSLRQVGLIGAFIYTLASFLAIFVVSTTQLIVTNGFLQGLGMGLLIPVSYTSFNSYFTKKKVLYLSLCKASIGLITMLYPLFIKFTITQYGFRGTLAIICAISAHSIFGALVMHPVQWYMVKKLKTCEKIVLIPPTPAIEAGKEKFHFNSNTTASKGAENARSVENLYAIKRTGSQKPTELLFVPKLADSRRLSIPVVLIPGETKADSKFNSSDNVYLENLYKAASVSSLGNFGNIAAEPMPVIKNKEGKWQTIMEYLDLTLLKDLVYDNIIVGMALTFFADLTFFTLEPLFLDKVNLNRSQIANIIALGGATDMCARLFLGVAGQFFIMNSRYMFFIGALFAAAFRLVMIQFTTYLPLLILTGILGALRSLVHIAQPIVMAEHVPIELYPSAYGLYMLLAGAISLSVGPMIGFIRDFTGSYVVAFVMLAVCNLCCVVPWAIEFIFRYKKERKLRNTDVS
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
A0A2A4JEJ5
A0A194PNS1
A0A2H1VXL3
A0A2W1BJL7
A0A182F9C7
B0XIY5
+ More
W5JLX9 Q17DF9 A0A182GDP8 A0A1Q3FJ07 A0A182MSX0 A0A182VGB7 A0A182RBY8 Q7QCE2 A0A182W0Y1 A0A182IPW6 A0A084WTD4 A0A182HSL8 A0A182TYU4 A0A1B0DHB5 A0A182NZV0 A0A182XAS2 A0A182NRI8 A0A182QJX4 A0A0M3QZ41 A0A182Y6H4 B4JK08 A0A1L8DEC6 A0A1B0B6K1 A0A1I8Q6B1 A0A0K8VXT5 A0A1A9ZB14 A0A182K474 B4M2A2 A0A1A9UZI4 A0A1A9W694 B3N140 A0A034WE28 A0A1L8EFW6 W8BP86 A0A1I8M485 A0A0Q5TAQ5 B4L850 A8JUR0 A0A0A1WGI3 B4PZJ4 B4NDJ4 B4I741 Q29GP4 A0A3B0KTR2 A0A1W4VFI3 A0A1L8EAR5 A0A2A4JNU3 B4GVD9 Q9VWJ2 A0A1B0GAB3 A0A2P8YB13 A0A1B0GJR3 A0A1L8DDT9 A0A212FNW6 A0A2P8YB42 A0A2H1VVT2 A0A088ABJ3 T1E9I8 A0A0L7R3L5 A0A0M8ZUQ5 A0A1Y1N8V7 A0A3L8DD75 A0A310SRY4 A0A0T6AZG7 A0A232FCW3 A0A1W4XGX8 K7J4F7 A0A212F801 A0A1W4WYB4 A0A1B6C167 A0A154PKP1 D2A1Q7 A0A1I8PQM0 A0A2A4K764 E2BFX0 A0A158NAT3 D2A1Q6 A0A1Y1LYE8 A0A194PVC1 A0A0A1XFI4 T1PG59 A0A1Q3G1R7 A0A1Q3G203 A0A2P8YB28 A0A2W1BKS8 A0A2R7X361 B4NUQ6 U4UMS7
W5JLX9 Q17DF9 A0A182GDP8 A0A1Q3FJ07 A0A182MSX0 A0A182VGB7 A0A182RBY8 Q7QCE2 A0A182W0Y1 A0A182IPW6 A0A084WTD4 A0A182HSL8 A0A182TYU4 A0A1B0DHB5 A0A182NZV0 A0A182XAS2 A0A182NRI8 A0A182QJX4 A0A0M3QZ41 A0A182Y6H4 B4JK08 A0A1L8DEC6 A0A1B0B6K1 A0A1I8Q6B1 A0A0K8VXT5 A0A1A9ZB14 A0A182K474 B4M2A2 A0A1A9UZI4 A0A1A9W694 B3N140 A0A034WE28 A0A1L8EFW6 W8BP86 A0A1I8M485 A0A0Q5TAQ5 B4L850 A8JUR0 A0A0A1WGI3 B4PZJ4 B4NDJ4 B4I741 Q29GP4 A0A3B0KTR2 A0A1W4VFI3 A0A1L8EAR5 A0A2A4JNU3 B4GVD9 Q9VWJ2 A0A1B0GAB3 A0A2P8YB13 A0A1B0GJR3 A0A1L8DDT9 A0A212FNW6 A0A2P8YB42 A0A2H1VVT2 A0A088ABJ3 T1E9I8 A0A0L7R3L5 A0A0M8ZUQ5 A0A1Y1N8V7 A0A3L8DD75 A0A310SRY4 A0A0T6AZG7 A0A232FCW3 A0A1W4XGX8 K7J4F7 A0A212F801 A0A1W4WYB4 A0A1B6C167 A0A154PKP1 D2A1Q7 A0A1I8PQM0 A0A2A4K764 E2BFX0 A0A158NAT3 D2A1Q6 A0A1Y1LYE8 A0A194PVC1 A0A0A1XFI4 T1PG59 A0A1Q3G1R7 A0A1Q3G203 A0A2P8YB28 A0A2W1BKS8 A0A2R7X361 B4NUQ6 U4UMS7
Pubmed
26354079
28756777
20920257
23761445
17510324
26483478
+ More
12364791 14747013 17210077 24438588 25244985 17994087 25348373 24495485 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25830018 17550304 15632085 29403074 22118469 28004739 30249741 28648823 20075255 18362917 19820115 20798317 21347285 23537049
12364791 14747013 17210077 24438588 25244985 17994087 25348373 24495485 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25830018 17550304 15632085 29403074 22118469 28004739 30249741 28648823 20075255 18362917 19820115 20798317 21347285 23537049
EMBL
NWSH01001768
PCG70196.1
KQ459597
KPI95076.1
ODYU01004743
SOQ44944.1
+ More
KZ150104 PZC73457.1 DS233409 EDS29793.1 ADMH02001190 ETN63784.1 CH477295 EAT44425.1 JXUM01010240 KQ560302 KXJ83164.1 GFDL01007537 JAV27508.1 AXCM01000202 AAAB01008859 EAA08073.5 ATLV01026883 KE525420 KFB53478.1 APCN01000295 AJVK01060889 AXCN02001457 CP012528 ALC48709.1 CH916370 EDV99910.1 GFDF01009272 JAV04812.1 JXJN01009134 GDHF01008606 JAI43708.1 CH940651 EDW65806.2 CH902650 EDV30075.2 GAKP01006008 JAC52944.1 GFDG01001263 JAV17536.1 GAMC01005833 JAC00723.1 CH954180 KQS30081.1 CH933814 EDW05625.1 AE014298 ABW09455.2 GBXI01016687 JAC97604.1 CM000162 EDX02150.2 CH964239 EDW82900.1 CH480823 EDW56139.1 CH379064 EAL32064.2 OUUW01000015 SPP88661.1 GFDG01002962 JAV15837.1 NWSH01001018 PCG73092.1 CH479192 EDW26426.1 AAF48946.3 CCAG010021887 PYGN01000741 PSN41452.1 AJWK01022144 AJWK01022145 AJWK01022146 GFDF01009468 JAV04616.1 AGBW02004200 OWR55438.1 PSN41453.1 SOQ44945.1 GAMD01001307 JAB00284.1 KQ414663 KOC65428.1 KQ435878 KOX69909.1 GEZM01012259 JAV93035.1 QOIP01000010 RLU18112.1 KQ761046 OAD58324.1 LJIG01022462 KRT80450.1 NNAY01000453 OXU28289.1 AGBW02009819 OWR49848.1 GEDC01030051 JAS07247.1 KQ434946 KZC12317.1 KQ971338 EFA01508.1 NWSH01000106 PCG79492.1 GL448096 EFN85372.1 ADTU01010093 EFA02707.1 GEZM01043968 GEZM01043967 GEZM01043966 JAV78624.1 KPI95075.1 GBXI01012470 GBXI01009867 GBXI01005049 GBXI01001190 JAD01822.1 JAD04425.1 JAD09243.1 JAD13102.1 KA647669 AFP62298.1 GFDL01001303 JAV33742.1 GFDL01001222 JAV33823.1 PSN41451.1 PZC73456.1 KK856674 PTY26239.1 CH984177 EDX16703.1 KB632374 ERL93798.1
KZ150104 PZC73457.1 DS233409 EDS29793.1 ADMH02001190 ETN63784.1 CH477295 EAT44425.1 JXUM01010240 KQ560302 KXJ83164.1 GFDL01007537 JAV27508.1 AXCM01000202 AAAB01008859 EAA08073.5 ATLV01026883 KE525420 KFB53478.1 APCN01000295 AJVK01060889 AXCN02001457 CP012528 ALC48709.1 CH916370 EDV99910.1 GFDF01009272 JAV04812.1 JXJN01009134 GDHF01008606 JAI43708.1 CH940651 EDW65806.2 CH902650 EDV30075.2 GAKP01006008 JAC52944.1 GFDG01001263 JAV17536.1 GAMC01005833 JAC00723.1 CH954180 KQS30081.1 CH933814 EDW05625.1 AE014298 ABW09455.2 GBXI01016687 JAC97604.1 CM000162 EDX02150.2 CH964239 EDW82900.1 CH480823 EDW56139.1 CH379064 EAL32064.2 OUUW01000015 SPP88661.1 GFDG01002962 JAV15837.1 NWSH01001018 PCG73092.1 CH479192 EDW26426.1 AAF48946.3 CCAG010021887 PYGN01000741 PSN41452.1 AJWK01022144 AJWK01022145 AJWK01022146 GFDF01009468 JAV04616.1 AGBW02004200 OWR55438.1 PSN41453.1 SOQ44945.1 GAMD01001307 JAB00284.1 KQ414663 KOC65428.1 KQ435878 KOX69909.1 GEZM01012259 JAV93035.1 QOIP01000010 RLU18112.1 KQ761046 OAD58324.1 LJIG01022462 KRT80450.1 NNAY01000453 OXU28289.1 AGBW02009819 OWR49848.1 GEDC01030051 JAS07247.1 KQ434946 KZC12317.1 KQ971338 EFA01508.1 NWSH01000106 PCG79492.1 GL448096 EFN85372.1 ADTU01010093 EFA02707.1 GEZM01043968 GEZM01043967 GEZM01043966 JAV78624.1 KPI95075.1 GBXI01012470 GBXI01009867 GBXI01005049 GBXI01001190 JAD01822.1 JAD04425.1 JAD09243.1 JAD13102.1 KA647669 AFP62298.1 GFDL01001303 JAV33742.1 GFDL01001222 JAV33823.1 PSN41451.1 PZC73456.1 KK856674 PTY26239.1 CH984177 EDX16703.1 KB632374 ERL93798.1
Proteomes
UP000218220
UP000053268
UP000069272
UP000002320
UP000000673
UP000008820
+ More
UP000069940 UP000249989 UP000075883 UP000075903 UP000075900 UP000007062 UP000075920 UP000075880 UP000030765 UP000075840 UP000075902 UP000092462 UP000075885 UP000076407 UP000075884 UP000075886 UP000092553 UP000076408 UP000001070 UP000092460 UP000095300 UP000092445 UP000075881 UP000008792 UP000078200 UP000091820 UP000007801 UP000095301 UP000008711 UP000009192 UP000000803 UP000002282 UP000007798 UP000001292 UP000001819 UP000268350 UP000192221 UP000008744 UP000092444 UP000245037 UP000092461 UP000007151 UP000005203 UP000053825 UP000053105 UP000279307 UP000215335 UP000192223 UP000002358 UP000076502 UP000007266 UP000008237 UP000005205 UP000000304 UP000030742
UP000069940 UP000249989 UP000075883 UP000075903 UP000075900 UP000007062 UP000075920 UP000075880 UP000030765 UP000075840 UP000075902 UP000092462 UP000075885 UP000076407 UP000075884 UP000075886 UP000092553 UP000076408 UP000001070 UP000092460 UP000095300 UP000092445 UP000075881 UP000008792 UP000078200 UP000091820 UP000007801 UP000095301 UP000008711 UP000009192 UP000000803 UP000002282 UP000007798 UP000001292 UP000001819 UP000268350 UP000192221 UP000008744 UP000092444 UP000245037 UP000092461 UP000007151 UP000005203 UP000053825 UP000053105 UP000279307 UP000215335 UP000192223 UP000002358 UP000076502 UP000007266 UP000008237 UP000005205 UP000000304 UP000030742
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JEJ5
A0A194PNS1
A0A2H1VXL3
A0A2W1BJL7
A0A182F9C7
B0XIY5
+ More
W5JLX9 Q17DF9 A0A182GDP8 A0A1Q3FJ07 A0A182MSX0 A0A182VGB7 A0A182RBY8 Q7QCE2 A0A182W0Y1 A0A182IPW6 A0A084WTD4 A0A182HSL8 A0A182TYU4 A0A1B0DHB5 A0A182NZV0 A0A182XAS2 A0A182NRI8 A0A182QJX4 A0A0M3QZ41 A0A182Y6H4 B4JK08 A0A1L8DEC6 A0A1B0B6K1 A0A1I8Q6B1 A0A0K8VXT5 A0A1A9ZB14 A0A182K474 B4M2A2 A0A1A9UZI4 A0A1A9W694 B3N140 A0A034WE28 A0A1L8EFW6 W8BP86 A0A1I8M485 A0A0Q5TAQ5 B4L850 A8JUR0 A0A0A1WGI3 B4PZJ4 B4NDJ4 B4I741 Q29GP4 A0A3B0KTR2 A0A1W4VFI3 A0A1L8EAR5 A0A2A4JNU3 B4GVD9 Q9VWJ2 A0A1B0GAB3 A0A2P8YB13 A0A1B0GJR3 A0A1L8DDT9 A0A212FNW6 A0A2P8YB42 A0A2H1VVT2 A0A088ABJ3 T1E9I8 A0A0L7R3L5 A0A0M8ZUQ5 A0A1Y1N8V7 A0A3L8DD75 A0A310SRY4 A0A0T6AZG7 A0A232FCW3 A0A1W4XGX8 K7J4F7 A0A212F801 A0A1W4WYB4 A0A1B6C167 A0A154PKP1 D2A1Q7 A0A1I8PQM0 A0A2A4K764 E2BFX0 A0A158NAT3 D2A1Q6 A0A1Y1LYE8 A0A194PVC1 A0A0A1XFI4 T1PG59 A0A1Q3G1R7 A0A1Q3G203 A0A2P8YB28 A0A2W1BKS8 A0A2R7X361 B4NUQ6 U4UMS7
W5JLX9 Q17DF9 A0A182GDP8 A0A1Q3FJ07 A0A182MSX0 A0A182VGB7 A0A182RBY8 Q7QCE2 A0A182W0Y1 A0A182IPW6 A0A084WTD4 A0A182HSL8 A0A182TYU4 A0A1B0DHB5 A0A182NZV0 A0A182XAS2 A0A182NRI8 A0A182QJX4 A0A0M3QZ41 A0A182Y6H4 B4JK08 A0A1L8DEC6 A0A1B0B6K1 A0A1I8Q6B1 A0A0K8VXT5 A0A1A9ZB14 A0A182K474 B4M2A2 A0A1A9UZI4 A0A1A9W694 B3N140 A0A034WE28 A0A1L8EFW6 W8BP86 A0A1I8M485 A0A0Q5TAQ5 B4L850 A8JUR0 A0A0A1WGI3 B4PZJ4 B4NDJ4 B4I741 Q29GP4 A0A3B0KTR2 A0A1W4VFI3 A0A1L8EAR5 A0A2A4JNU3 B4GVD9 Q9VWJ2 A0A1B0GAB3 A0A2P8YB13 A0A1B0GJR3 A0A1L8DDT9 A0A212FNW6 A0A2P8YB42 A0A2H1VVT2 A0A088ABJ3 T1E9I8 A0A0L7R3L5 A0A0M8ZUQ5 A0A1Y1N8V7 A0A3L8DD75 A0A310SRY4 A0A0T6AZG7 A0A232FCW3 A0A1W4XGX8 K7J4F7 A0A212F801 A0A1W4WYB4 A0A1B6C167 A0A154PKP1 D2A1Q7 A0A1I8PQM0 A0A2A4K764 E2BFX0 A0A158NAT3 D2A1Q6 A0A1Y1LYE8 A0A194PVC1 A0A0A1XFI4 T1PG59 A0A1Q3G1R7 A0A1Q3G203 A0A2P8YB28 A0A2W1BKS8 A0A2R7X361 B4NUQ6 U4UMS7
Ontologies
GO
Topology
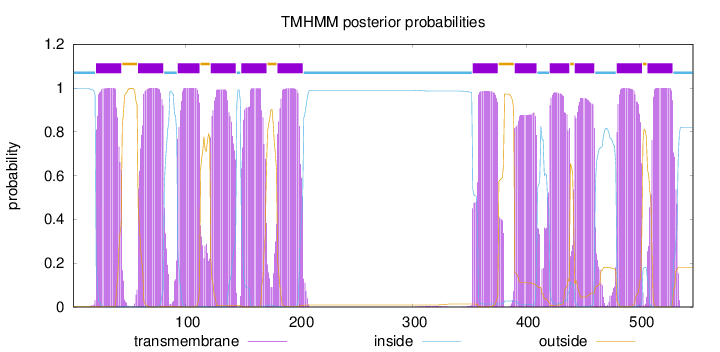
Length:
547
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
255.90336
Exp number, first 60 AAs:
23.86175
Total prob of N-in:
0.99788
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 57
TMhelix
58 - 80
inside
81 - 92
TMhelix
93 - 112
outside
113 - 121
TMhelix
122 - 144
inside
145 - 148
TMhelix
149 - 171
outside
172 - 180
TMhelix
181 - 203
inside
204 - 352
TMhelix
353 - 375
outside
376 - 389
TMhelix
390 - 409
inside
410 - 420
TMhelix
421 - 438
outside
439 - 442
TMhelix
443 - 460
inside
461 - 479
TMhelix
480 - 502
outside
503 - 506
TMhelix
507 - 529
inside
530 - 547
Population Genetic Test Statistics
Pi
211.858097
Theta
162.430811
Tajima's D
0.735883
CLR
0.522333
CSRT
0.590670466476676
Interpretation
Uncertain
