Gene
KWMTBOMO02957
Pre Gene Modal
BGIBMGA003568
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_trypsin?_alkaline_C-like_[Bombyx_mori]
Full name
Trypsin, alkaline A
+ More
Trypsin, alkaline C
Trypsin, alkaline B
Trypsin CFT-1
Trypsin, alkaline C
Trypsin, alkaline B
Trypsin CFT-1
Location in the cell
Extracellular Reliability : 4.578
Sequence
CDS
ATGACACCGACGCTCGTCTTACTGCTCTTAGTTGGAGCTGTGGCTGCCATACCAGACCAGTCCTCAAGGATTGTCGGTGGGACAGCGACAACTATCGACCGGTACCCCACAATGGCAGCAGCACTTTTTTCTAGCAATATGGCCCAGTGGTGGCAAGGATGTGGCTGTTCCATTCTCAATAACAGAGCCGTCCTTACAGCCGCACATTGTATTATAGGAGATCGTGTCGGTGTTTGGAGAGTTCGCGTAGGATCTTCTTTTGGAAATAGTGGAGGTGTTGTCCATGAGGTCAACAGGCACATCATCCACCCTAATTACAATCATCGTACAAAGGACGCTGATATTGCAATCATGCGTACAGTTTTACGGATAAACTTCGTACGTAATACCGTGCAGCCAGCTCGTATTGCGGGTTCCAATTACAACCTTGCCGATAACCAAGTTGTTTGGGCAGCTGGATGGGGATCTACAAGGACTGGAAGTGGTTTTGCTGAACAGCTTCGACATGTTCAAGTTTGGACGATAAATCAGGCCATTTGTAGAGAGCGATACTCGAGAATAGATAGACCGATCACTGCCAATATGTTGTGCTCCGGCTGGCTTGACGTCGGCGGCCGCGATCAGTGTGGAGGTGATTCCGGTGGTCCTCTATACCACAACGGTGTCGTCGTCGGGGTGTGCTCGTGGGGATTCTCTTGTGGAAGTGCATTCTACCCTGGAGTAAATGTTCGCGTCTCTCGCTTCAGTTCATGGATTCAAAACAATGCTTAG
Protein
MTPTLVLLLLVGAVAAIPDQSSRIVGGTATTIDRYPTMAAALFSSNMAQWWQGCGCSILNNRAVLTAAHCIIGDRVGVWRVRVGSSFGNSGGVVHEVNRHIIHPNYNHRTKDADIAIMRTVLRINFVRNTVQPARIAGSNYNLADNQVVWAAGWGSTRTGSGFAEQLRHVQVWTINQAICRERYSRIDRPITANMLCSGWLDVGGRDQCGGDSGGPLYHNGVVVGVCSWGFSCGSAFYPGVNVRVSRFSSWIQNNA
Summary
Catalytic Activity
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Disulfide bond
Hydrolase
Protease
Secreted
Serine protease
Signal
Zymogen
Direct protein sequencing
Feature
propeptide Activation peptide
chain Trypsin, alkaline A
chain Trypsin, alkaline A
Uniprot
H9J231
P35045
P35047
Q56IB6
P35046
Q56IB5
+ More
Q56IB8 Q56IB4 Q6R561 Q56IB7 Q1HQA7 I6TMW8 H9J229 J7JYV8 A0A2A4K6F3 H9J232 Q9TXE6 A0A2A4K5F8 H9J230 A0A2W1BKV8 J7IJ83 V9M0R7 B4X995 A0A2A4K5G7 A0A2A4K6E9 A0A2A4K6E3 A0A194PNR6 Q27540 I3UII8 B4X996 P35042 A0A2A4K6W3 O18447 I7DEI4 A0A2H1VJ41 A0A2A4K648 A0A2W1BSA6 A0A194PVB6 A0A2W1BQW9 A0A194PQQ0 A0A1C9M3A1 A0A194PQQ5 B4X9A1 W5X3N9 A0A194PNR3 Q961Y0 J7IF09 A0A2A4K6U2 J7IFG7 T1QDI0 E7D004 A0A2W1B606 A0A0L7KUC9 A0A2W1BA14 I7E449 O18434 A0A2A4K747 O18436 A0A1B0RHN7 A0A2W1BGQ7 A0A2W1BF78 A0A194PP33 S5PVP1 A0A0K8SII3 A0A2H1V0W3 C4PFX2 C9W8F7 A0A2A4K769 A0A2W1BP39 B3F884 O76954 A0A2U8NFD7 A0A0N1PHJ0 A0A2W1B1G1 A0A2W1BKF9 A0A2A4K5I4 Q4L1K4 D7S002 A9P3S8 H9J243 B4X9A0 V9LL27 O18441 D7S000 A0A2A4K6F8 A0A2W1BE44 A0A2W1BL45 O62598 O18435 Q4L1K6 A0A345H7U4 D7S001 A0A2H1W4N7 A0A0N1I947 A0A2A4K5I0 A0A2W1BKR8 A0A2W1BGR2 A0A2W1BP46 I3UII6 A0A1E1WNA7
Q56IB8 Q56IB4 Q6R561 Q56IB7 Q1HQA7 I6TMW8 H9J229 J7JYV8 A0A2A4K6F3 H9J232 Q9TXE6 A0A2A4K5F8 H9J230 A0A2W1BKV8 J7IJ83 V9M0R7 B4X995 A0A2A4K5G7 A0A2A4K6E9 A0A2A4K6E3 A0A194PNR6 Q27540 I3UII8 B4X996 P35042 A0A2A4K6W3 O18447 I7DEI4 A0A2H1VJ41 A0A2A4K648 A0A2W1BSA6 A0A194PVB6 A0A2W1BQW9 A0A194PQQ0 A0A1C9M3A1 A0A194PQQ5 B4X9A1 W5X3N9 A0A194PNR3 Q961Y0 J7IF09 A0A2A4K6U2 J7IFG7 T1QDI0 E7D004 A0A2W1B606 A0A0L7KUC9 A0A2W1BA14 I7E449 O18434 A0A2A4K747 O18436 A0A1B0RHN7 A0A2W1BGQ7 A0A2W1BF78 A0A194PP33 S5PVP1 A0A0K8SII3 A0A2H1V0W3 C4PFX2 C9W8F7 A0A2A4K769 A0A2W1BP39 B3F884 O76954 A0A2U8NFD7 A0A0N1PHJ0 A0A2W1B1G1 A0A2W1BKF9 A0A2A4K5I4 Q4L1K4 D7S002 A9P3S8 H9J243 B4X9A0 V9LL27 O18441 D7S000 A0A2A4K6F8 A0A2W1BE44 A0A2W1BL45 O62598 O18435 Q4L1K6 A0A345H7U4 D7S001 A0A2H1W4N7 A0A0N1I947 A0A2A4K5I0 A0A2W1BKR8 A0A2W1BGR2 A0A2W1BP46 I3UII6 A0A1E1WNA7
EC Number
3.4.21.4
Pubmed
EMBL
BABH01007514
BABH01007515
L16805
L16807
AY953061
AAX62034.1
+ More
L16806 AY953062 AAX62035.1 AY953059 AAX62032.1 AY953063 AAX62036.1 AY513649 AAR98918.1 AY953060 AAX62033.1 DQ443145 ABF51234.1 JQ904123 AFM77753.1 BABH01007520 JX312360 AFQ59994.1 NWSH01000106 PCG79484.1 PCG79475.1 BABH01007517 KZ149992 PZC75528.1 JX195651 AFQ37934.1 JX866720 AGB93875.1 EF531631 EF531633 ABR88242.1 ABR88244.1 PCG79485.1 PCG79488.1 PCG79474.1 KQ459597 KPI95071.1 U12917 AAA84423.1 JN315762 AFK64826.1 EF531632 ABR88243.1 L04749 PCG79493.1 HM209426 Y12283 KZ150104 ADI32887.1 CAA72962.1 PZC73444.1 JN793539 AFO68319.1 ODYU01002655 SOQ40442.1 PCG79486.1 PZC75526.1 KPI95070.1 PZC75527.1 KPI95069.1 KU302250 AOQ30448.1 KPI95074.1 EF531637 ABR88248.1 KF779933 AHI07442.1 KPI95066.1 AY040819 AAK81696.1 JX195649 AFQ37932.1 PCG79473.1 JX195652 AFQ37935.1 JQ429438 AFW03963.1 HM990174 ADT80822.1 KZ150747 PZC70335.1 JTDY01005557 KOB66887.1 PZC70337.1 JX046916 AFO83992.1 Y12269 CAA72948.1 PZC73449.1 PCG79472.1 EF104916 Y12271 ABP96918.1 CAA72950.1 KM360188 AKH49604.1 PZC73441.1 PZC73448.1 KPI95067.1 KC175562 AGR92345.1 GBRD01013231 JAG52595.1 ODYU01000133 SOQ34396.1 FJ940726 ODYU01006287 ACR25157.1 SOQ47996.1 FJ205402 ACR15970.1 PCG79490.1 PZC73443.1 EF635223 ABU96714.1 AJ007706 CAA07611.1 MF407317 AWL83213.1 KQ460226 KPJ16473.1 PZC70338.1 PZC75529.1 PCG79487.1 AY587161 AAT95353.1 HM209425 ADI32886.1 EF104917 ABP96919.1 BABH01007473 EF531636 ABR88247.1 PCG79483.1 JQ340915 AFP74111.1 Y12276 CAA72955.1 PZC70339.1 HM209423 ADI32884.1 PCG79498.1 PZC73442.1 PZC73450.1 AF064525 AF064526 AAC36247.1 AAC36248.1 Y12270 CAA72949.1 AY587159 AAT95351.1 MF770251 AXG72654.1 HM209424 ADI32885.1 ODYU01007611 SOQ47997.1 SOQ50538.1 KPJ16474.1 PCG79481.1 PZC73446.1 PZC73451.1 PZC73453.1 JN315760 AFK64824.1 GDQN01002626 JAT88428.1
L16806 AY953062 AAX62035.1 AY953059 AAX62032.1 AY953063 AAX62036.1 AY513649 AAR98918.1 AY953060 AAX62033.1 DQ443145 ABF51234.1 JQ904123 AFM77753.1 BABH01007520 JX312360 AFQ59994.1 NWSH01000106 PCG79484.1 PCG79475.1 BABH01007517 KZ149992 PZC75528.1 JX195651 AFQ37934.1 JX866720 AGB93875.1 EF531631 EF531633 ABR88242.1 ABR88244.1 PCG79485.1 PCG79488.1 PCG79474.1 KQ459597 KPI95071.1 U12917 AAA84423.1 JN315762 AFK64826.1 EF531632 ABR88243.1 L04749 PCG79493.1 HM209426 Y12283 KZ150104 ADI32887.1 CAA72962.1 PZC73444.1 JN793539 AFO68319.1 ODYU01002655 SOQ40442.1 PCG79486.1 PZC75526.1 KPI95070.1 PZC75527.1 KPI95069.1 KU302250 AOQ30448.1 KPI95074.1 EF531637 ABR88248.1 KF779933 AHI07442.1 KPI95066.1 AY040819 AAK81696.1 JX195649 AFQ37932.1 PCG79473.1 JX195652 AFQ37935.1 JQ429438 AFW03963.1 HM990174 ADT80822.1 KZ150747 PZC70335.1 JTDY01005557 KOB66887.1 PZC70337.1 JX046916 AFO83992.1 Y12269 CAA72948.1 PZC73449.1 PCG79472.1 EF104916 Y12271 ABP96918.1 CAA72950.1 KM360188 AKH49604.1 PZC73441.1 PZC73448.1 KPI95067.1 KC175562 AGR92345.1 GBRD01013231 JAG52595.1 ODYU01000133 SOQ34396.1 FJ940726 ODYU01006287 ACR25157.1 SOQ47996.1 FJ205402 ACR15970.1 PCG79490.1 PZC73443.1 EF635223 ABU96714.1 AJ007706 CAA07611.1 MF407317 AWL83213.1 KQ460226 KPJ16473.1 PZC70338.1 PZC75529.1 PCG79487.1 AY587161 AAT95353.1 HM209425 ADI32886.1 EF104917 ABP96919.1 BABH01007473 EF531636 ABR88247.1 PCG79483.1 JQ340915 AFP74111.1 Y12276 CAA72955.1 PZC70339.1 HM209423 ADI32884.1 PCG79498.1 PZC73442.1 PZC73450.1 AF064525 AF064526 AAC36247.1 AAC36248.1 Y12270 CAA72949.1 AY587159 AAT95351.1 MF770251 AXG72654.1 HM209424 ADI32885.1 ODYU01007611 SOQ47997.1 SOQ50538.1 KPJ16474.1 PCG79481.1 PZC73446.1 PZC73451.1 PZC73453.1 JN315760 AFK64824.1 GDQN01002626 JAT88428.1
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9J231
P35045
P35047
Q56IB6
P35046
Q56IB5
+ More
Q56IB8 Q56IB4 Q6R561 Q56IB7 Q1HQA7 I6TMW8 H9J229 J7JYV8 A0A2A4K6F3 H9J232 Q9TXE6 A0A2A4K5F8 H9J230 A0A2W1BKV8 J7IJ83 V9M0R7 B4X995 A0A2A4K5G7 A0A2A4K6E9 A0A2A4K6E3 A0A194PNR6 Q27540 I3UII8 B4X996 P35042 A0A2A4K6W3 O18447 I7DEI4 A0A2H1VJ41 A0A2A4K648 A0A2W1BSA6 A0A194PVB6 A0A2W1BQW9 A0A194PQQ0 A0A1C9M3A1 A0A194PQQ5 B4X9A1 W5X3N9 A0A194PNR3 Q961Y0 J7IF09 A0A2A4K6U2 J7IFG7 T1QDI0 E7D004 A0A2W1B606 A0A0L7KUC9 A0A2W1BA14 I7E449 O18434 A0A2A4K747 O18436 A0A1B0RHN7 A0A2W1BGQ7 A0A2W1BF78 A0A194PP33 S5PVP1 A0A0K8SII3 A0A2H1V0W3 C4PFX2 C9W8F7 A0A2A4K769 A0A2W1BP39 B3F884 O76954 A0A2U8NFD7 A0A0N1PHJ0 A0A2W1B1G1 A0A2W1BKF9 A0A2A4K5I4 Q4L1K4 D7S002 A9P3S8 H9J243 B4X9A0 V9LL27 O18441 D7S000 A0A2A4K6F8 A0A2W1BE44 A0A2W1BL45 O62598 O18435 Q4L1K6 A0A345H7U4 D7S001 A0A2H1W4N7 A0A0N1I947 A0A2A4K5I0 A0A2W1BKR8 A0A2W1BGR2 A0A2W1BP46 I3UII6 A0A1E1WNA7
Q56IB8 Q56IB4 Q6R561 Q56IB7 Q1HQA7 I6TMW8 H9J229 J7JYV8 A0A2A4K6F3 H9J232 Q9TXE6 A0A2A4K5F8 H9J230 A0A2W1BKV8 J7IJ83 V9M0R7 B4X995 A0A2A4K5G7 A0A2A4K6E9 A0A2A4K6E3 A0A194PNR6 Q27540 I3UII8 B4X996 P35042 A0A2A4K6W3 O18447 I7DEI4 A0A2H1VJ41 A0A2A4K648 A0A2W1BSA6 A0A194PVB6 A0A2W1BQW9 A0A194PQQ0 A0A1C9M3A1 A0A194PQQ5 B4X9A1 W5X3N9 A0A194PNR3 Q961Y0 J7IF09 A0A2A4K6U2 J7IFG7 T1QDI0 E7D004 A0A2W1B606 A0A0L7KUC9 A0A2W1BA14 I7E449 O18434 A0A2A4K747 O18436 A0A1B0RHN7 A0A2W1BGQ7 A0A2W1BF78 A0A194PP33 S5PVP1 A0A0K8SII3 A0A2H1V0W3 C4PFX2 C9W8F7 A0A2A4K769 A0A2W1BP39 B3F884 O76954 A0A2U8NFD7 A0A0N1PHJ0 A0A2W1B1G1 A0A2W1BKF9 A0A2A4K5I4 Q4L1K4 D7S002 A9P3S8 H9J243 B4X9A0 V9LL27 O18441 D7S000 A0A2A4K6F8 A0A2W1BE44 A0A2W1BL45 O62598 O18435 Q4L1K6 A0A345H7U4 D7S001 A0A2H1W4N7 A0A0N1I947 A0A2A4K5I0 A0A2W1BKR8 A0A2W1BGR2 A0A2W1BP46 I3UII6 A0A1E1WNA7
PDB
3W94
E-value=1.11563e-15,
Score=201
Ontologies
GO
Topology
Subcellular location
SignalP
Position: 1 - 16,
Likelihood: 0.993118
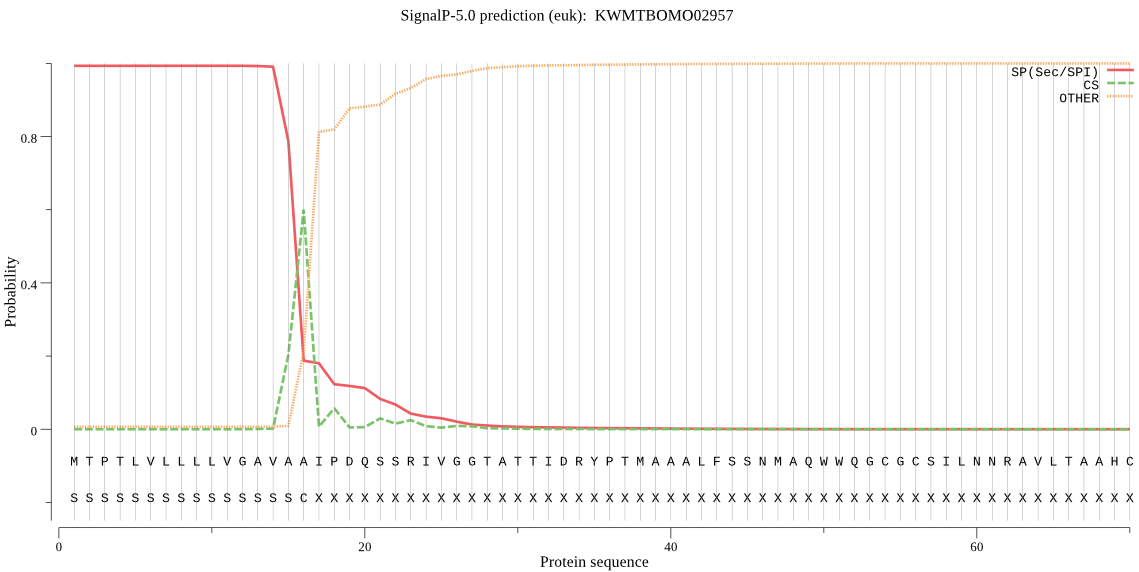
Length:
256
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.4131
Exp number, first 60 AAs:
0.31354
Total prob of N-in:
0.01288
outside
1 - 256

Population Genetic Test Statistics
Pi
163.867894
Theta
142.288582
Tajima's D
0.453834
CLR
0.137013
CSRT
0.497825108744563
Interpretation
Uncertain
