Gene
KWMTBOMO02949
Pre Gene Modal
BGIBMGA003629
Annotation
PREDICTED:_solute_carrier_family_12_member_4_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.93
Sequence
CDS
ATGGAATCAGAAACTTGTTCCTGTTTCGACTCCGTATGTGTATATAAAGAAGGTGACCCCGACGAACATGGACTATCCACGGACAAGAATAAAGGCTGCGACACCAACCTTTATCTCTATCACGAAGAGATCGAGGACCGACCTAGAGCAGCCACGTTCCTCAGCTCACTAGCGGACTATGCGAATACAATCCCCACGGCGTCCGCCGCAGACCCTGATGCCCCCAAGCCAGCCCCACCAGCCCGCATGGGCACGCTCATCGGCGTGTACCTGCCTTGCATCCAGAATATCTTCGGTGTGATCCTATTCATCCGACTCACCTGGGTTGTCGGCACCGCTGGCGCTATACAAGGATTCCTCATTGTTCTAACCTGCTGCTGCACGACGATGTTAACCGCAATATCAATGTCTGCCATCGCAACGAACGGCGTGGTGCCGGCGGGGGGATCGTATTTCATGATCGGTCGGTCCCTGGGGCCGGAGTGCGGCGGGGCGGTCGGCATGCTGTTCTACACGGGTACAACCCTAGCGGCCGCTATGTACATCGTTGGAGCTGTCGAAATCGTACTGACTTACATGGCACCGTGGATATCAATTTTCGGCGATTTCACTAAGGACCCAGAAGCAATGTACAACAATTTCCGCGTCTATGGAACCGGTTTGCTCCTCATTATGGGGATGGTTGTTTTCGTTGGAGTAAAGTTCGTCAACAAATTTGCTACTGTCGCTCTAGCCTGCGTCATCCTCTCCATCACCGCGGTATATGCCGGAATATTCGTCAACTTCTATGGAAACGATAGCTTACAAATGTGTGTACTCGGACGCCGCTTGCTCAAGGATATTCATATTAGCAATTGCACAAAGGACCCAAAAGGCGAACTGCATGCAATGTTCTGTCCCGATAAAATCTGTGACCCCTACTACGAAGCCCATAACATAAGCATTGTGCAAGGAATTAAGGGTCTCGCAAGTGGGGTGTTCTTCGATAACCTCCAGGACTCGTTCATCCAAGAAGGTCAATACATCGCATACGGCAAGGAACCTGACGACATTGAACAGATGGAGAGACCTACCCACAATCAAATTTACGCGGACATCACTACGACTTTCACAATTTTGATAGGCATATTCTTCCCTTCGGTGACCGGCATAATGGCTGGTTCAAATCGCTCCGGAGACTTGGCCGATGCTCAAAAGAGTATTCCAATTGGAACTATTTGTGCGATCCTCACCACTTCCACCGTCTACCTCTCGTGTGTGCTGTTATTTGCTGGCACTGTCGATAATTTGTTACTCAGAGATAAATTCGGTCAATCGATTGGCGGGAAATTAGTCGTTGCGAACATGGCGTGGCCAAACCAATGGGTGATTCTGATCGGCTCATTCCTGTCGACATTGGGCGCGGGACTCCAATCTCTAACCGGTGCGCCGCGACTACTGCAAGCCATCGCCAAAGATGAAATCATTCCTTTTCTCGCTCCATTCGCAGTCTCTTCCAAACGAGGAGAGCCTACCCGGGCCTTACTTCTGACCATGGCAATCTGCCAATGCGGCATCTTACTCGGGAACGTCGACATCTTGGCTCCCCTGTTGTCCATGTTCTTCTTAATGTGCTACGGATTCGTCAACTTGGCTTGTGCTCTTCAAACTCTACTCAAGACCCCGAACTGGCGTCCTCGCTTCAAATACTACCACTGGTAA
Protein
MESETCSCFDSVCVYKEGDPDEHGLSTDKNKGCDTNLYLYHEEIEDRPRAATFLSSLADYANTIPTASAADPDAPKPAPPARMGTLIGVYLPCIQNIFGVILFIRLTWVVGTAGAIQGFLIVLTCCCTTMLTAISMSAIATNGVVPAGGSYFMIGRSLGPECGGAVGMLFYTGTTLAAAMYIVGAVEIVLTYMAPWISIFGDFTKDPEAMYNNFRVYGTGLLLIMGMVVFVGVKFVNKFATVALACVILSITAVYAGIFVNFYGNDSLQMCVLGRRLLKDIHISNCTKDPKGELHAMFCPDKICDPYYEAHNISIVQGIKGLASGVFFDNLQDSFIQEGQYIAYGKEPDDIEQMERPTHNQIYADITTTFTILIGIFFPSVTGIMAGSNRSGDLADAQKSIPIGTICAILTTSTVYLSCVLLFAGTVDNLLLRDKFGQSIGGKLVVANMAWPNQWVILIGSFLSTLGAGLQSLTGAPRLLQAIAKDEIIPFLAPFAVSSKRGEPTRALLLTMAICQCGILLGNVDILAPLLSMFFLMCYGFVNLACALQTLLKTPNWRPRFKYYHW
Summary
Uniprot
H9J292
A0A0N0PDE9
A0A2A4K3L9
A0A2W1BKK4
A0A194PVB2
A0A2H1VVD7
+ More
A0A2Z4GWW8 A0A0L7LEL9 A0A2M3ZHQ8 A0A1Q3G591 A0A2M4AJX9 A0A1Q3G587 A0A1L8DEP3 A0A1Q3G595 A0A1I8JU73 A0A1B0GIR5 A0A1I8JT46 A0A1W4X8E4 A0A1C7CZA6 A0A1I8JVP3 A0A1I8JT60 A0A1Y9IHS4 A0A1I8JVP2 A0A1I8JVL0 A0A1W4X876 A0A1W4X6Z7 A0A1W4WXM6 H2L215 A0A023F5C7 Q16P15 A0A1I8JVE1 A0A1I8JVB6 A0A1I8JVB5 A0A0A9VUE7 A0A0K8S3J5 A0A0K8S552 A0A0K8S3X9 A0A146M9C9 A0A0A9W1U4 A0A087ZZW6 V9I9A6 V9IA86 V9I7X5 V9I9S9 A0A0L0C9C4 A0A2P8XHN8 T1PD66 A0A0Q9XIN2 A0A0Q9XKJ7 A0A1B6ES16 W8BPR2 A0A1B6G3Y2 B4KMC3 A0A0Q9XM85 A0A0Q9X8Z8 A0A158NKY1 W8C206 A0A1I8M4N6 A0A1I8M4N7 A0A0Q9XI19 T1PD35 A0A1I8M4N4 A0A1I8QA37 A0A034VI57 A0A1I8QA21 A0A0K8V8W7 A0A0K8UL13 A0A3B0JIV8 A0A1I8Q9X8 A0A034VJ83 A0A3B0IYU5 A0A034VKW0 A0A0K8W0Z0 A0A0P8YAI4 A0A1W4UYZ9 A0A154P501 A0A3B0J2E5 A0A3B0IYK7 A0A034VK61 A0A0P9BX49 B3MCM5 A0A0P8Y2U3 A0A0Q5VP73 A0A1W4UM20 A0A1W4UZA2 A0A1W4UZA7 A0A0J9RJF1 A0A0Q5VQ76 A0A1W4V0K7 V5G4T2 A0A0B4LGD3 A0A0P8XN64 A0A0R1DTU3 A0A1W4ULI7 A0A1W4V0K9 A0A0J9RL32 Q9W1G5 A0A0R1DTT7 A0A0N8P044 A0A0P8XN79
A0A2Z4GWW8 A0A0L7LEL9 A0A2M3ZHQ8 A0A1Q3G591 A0A2M4AJX9 A0A1Q3G587 A0A1L8DEP3 A0A1Q3G595 A0A1I8JU73 A0A1B0GIR5 A0A1I8JT46 A0A1W4X8E4 A0A1C7CZA6 A0A1I8JVP3 A0A1I8JT60 A0A1Y9IHS4 A0A1I8JVP2 A0A1I8JVL0 A0A1W4X876 A0A1W4X6Z7 A0A1W4WXM6 H2L215 A0A023F5C7 Q16P15 A0A1I8JVE1 A0A1I8JVB6 A0A1I8JVB5 A0A0A9VUE7 A0A0K8S3J5 A0A0K8S552 A0A0K8S3X9 A0A146M9C9 A0A0A9W1U4 A0A087ZZW6 V9I9A6 V9IA86 V9I7X5 V9I9S9 A0A0L0C9C4 A0A2P8XHN8 T1PD66 A0A0Q9XIN2 A0A0Q9XKJ7 A0A1B6ES16 W8BPR2 A0A1B6G3Y2 B4KMC3 A0A0Q9XM85 A0A0Q9X8Z8 A0A158NKY1 W8C206 A0A1I8M4N6 A0A1I8M4N7 A0A0Q9XI19 T1PD35 A0A1I8M4N4 A0A1I8QA37 A0A034VI57 A0A1I8QA21 A0A0K8V8W7 A0A0K8UL13 A0A3B0JIV8 A0A1I8Q9X8 A0A034VJ83 A0A3B0IYU5 A0A034VKW0 A0A0K8W0Z0 A0A0P8YAI4 A0A1W4UYZ9 A0A154P501 A0A3B0J2E5 A0A3B0IYK7 A0A034VK61 A0A0P9BX49 B3MCM5 A0A0P8Y2U3 A0A0Q5VP73 A0A1W4UM20 A0A1W4UZA2 A0A1W4UZA7 A0A0J9RJF1 A0A0Q5VQ76 A0A1W4V0K7 V5G4T2 A0A0B4LGD3 A0A0P8XN64 A0A0R1DTU3 A0A1W4ULI7 A0A1W4V0K9 A0A0J9RL32 Q9W1G5 A0A0R1DTT7 A0A0N8P044 A0A0P8XN79
Pubmed
EMBL
BABH01007527
BABH01007528
BABH01007529
KQ460226
KPJ16471.1
NWSH01000167
+ More
PCG78861.1 KZ149992 PZC75569.1 KQ459597 KPI95065.1 ODYU01004662 SOQ44790.1 MH048896 AWW06968.1 JTDY01001458 KOB73819.1 GGFM01007302 MBW28053.1 GFDL01000080 JAV34965.1 GGFK01007760 MBW41081.1 GFDL01000077 JAV34968.1 GFDF01009155 JAV04929.1 GFDL01000078 JAV34967.1 AJWK01015337 AJWK01015338 AJWK01015339 AJWK01015340 AJWK01015341 AJWK01015342 AJWK01015343 APCN01001926 APCN01001927 APCN01001928 APCN01001929 AAAB01008966 HM125960 ADM89630.1 GBBI01002394 JAC16318.1 CH477799 EAT36104.1 GBHO01044768 JAF98835.1 GBRD01017965 JAG47862.1 GBRD01017962 JAG47865.1 GBRD01017963 JAG47864.1 GDHC01002276 JAQ16353.1 GBHO01044769 JAF98834.1 JR036560 JR036561 AEY57226.1 AEY57227.1 JR036563 JR036564 AEY57229.1 AEY57230.1 JR036562 AEY57228.1 JR036565 AEY57231.1 JRES01000737 KNC28852.1 PYGN01002098 PSN31506.1 KA646701 AFP61330.1 CH933808 KRG04911.1 KRG04914.1 KRG04917.1 KRG04913.1 GECZ01028984 JAS40785.1 GAMC01003336 GAMC01003335 GAMC01003334 JAC03221.1 GECZ01012625 JAS57144.1 EDW09811.1 KRG04915.1 KRG04912.1 ADTU01019023 ADTU01019024 ADTU01019025 ADTU01019026 GAMC01003337 JAC03219.1 KRG04916.1 KA646702 AFP61331.1 GAKP01016783 JAC42169.1 GDHF01019988 GDHF01018775 GDHF01016925 GDHF01002908 JAI32326.1 JAI33539.1 JAI35389.1 JAI49406.1 GDHF01025264 GDHF01024059 GDHF01010894 GDHF01000286 JAI27050.1 JAI28255.1 JAI41420.1 JAI52028.1 OUUW01000001 SPP73176.1 GAKP01016785 GAKP01016780 JAC42167.1 SPP73175.1 GAKP01016784 JAC42168.1 GDHF01013653 GDHF01007568 JAI38661.1 JAI44746.1 CH902619 KPU76093.1 KQ434809 KZC06268.1 SPP73173.1 SPP73174.1 GAKP01016782 JAC42170.1 KPU76094.1 EDV36259.1 KPU76096.1 CH954179 KQS63069.1 KQS63075.1 CM002911 KMY96163.1 KQS63076.1 GALX01003421 JAB65045.1 AE013599 AHN56590.1 KPU76097.1 CM000158 KRK00502.1 KRK00509.1 KMY96164.1 AAF47099.2 KRK00501.1 KPU76092.1 KPU76091.1
PCG78861.1 KZ149992 PZC75569.1 KQ459597 KPI95065.1 ODYU01004662 SOQ44790.1 MH048896 AWW06968.1 JTDY01001458 KOB73819.1 GGFM01007302 MBW28053.1 GFDL01000080 JAV34965.1 GGFK01007760 MBW41081.1 GFDL01000077 JAV34968.1 GFDF01009155 JAV04929.1 GFDL01000078 JAV34967.1 AJWK01015337 AJWK01015338 AJWK01015339 AJWK01015340 AJWK01015341 AJWK01015342 AJWK01015343 APCN01001926 APCN01001927 APCN01001928 APCN01001929 AAAB01008966 HM125960 ADM89630.1 GBBI01002394 JAC16318.1 CH477799 EAT36104.1 GBHO01044768 JAF98835.1 GBRD01017965 JAG47862.1 GBRD01017962 JAG47865.1 GBRD01017963 JAG47864.1 GDHC01002276 JAQ16353.1 GBHO01044769 JAF98834.1 JR036560 JR036561 AEY57226.1 AEY57227.1 JR036563 JR036564 AEY57229.1 AEY57230.1 JR036562 AEY57228.1 JR036565 AEY57231.1 JRES01000737 KNC28852.1 PYGN01002098 PSN31506.1 KA646701 AFP61330.1 CH933808 KRG04911.1 KRG04914.1 KRG04917.1 KRG04913.1 GECZ01028984 JAS40785.1 GAMC01003336 GAMC01003335 GAMC01003334 JAC03221.1 GECZ01012625 JAS57144.1 EDW09811.1 KRG04915.1 KRG04912.1 ADTU01019023 ADTU01019024 ADTU01019025 ADTU01019026 GAMC01003337 JAC03219.1 KRG04916.1 KA646702 AFP61331.1 GAKP01016783 JAC42169.1 GDHF01019988 GDHF01018775 GDHF01016925 GDHF01002908 JAI32326.1 JAI33539.1 JAI35389.1 JAI49406.1 GDHF01025264 GDHF01024059 GDHF01010894 GDHF01000286 JAI27050.1 JAI28255.1 JAI41420.1 JAI52028.1 OUUW01000001 SPP73176.1 GAKP01016785 GAKP01016780 JAC42167.1 SPP73175.1 GAKP01016784 JAC42168.1 GDHF01013653 GDHF01007568 JAI38661.1 JAI44746.1 CH902619 KPU76093.1 KQ434809 KZC06268.1 SPP73173.1 SPP73174.1 GAKP01016782 JAC42170.1 KPU76094.1 EDV36259.1 KPU76096.1 CH954179 KQS63069.1 KQS63075.1 CM002911 KMY96163.1 KQS63076.1 GALX01003421 JAB65045.1 AE013599 AHN56590.1 KPU76097.1 CM000158 KRK00502.1 KRK00509.1 KMY96164.1 AAF47099.2 KRK00501.1 KPU76092.1 KPU76091.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000037510
UP000075900
+ More
UP000092461 UP000075840 UP000192223 UP000075882 UP000076407 UP000008820 UP000075903 UP000005203 UP000037069 UP000245037 UP000095301 UP000009192 UP000005205 UP000095300 UP000268350 UP000007801 UP000192221 UP000076502 UP000008711 UP000000803 UP000002282
UP000092461 UP000075840 UP000192223 UP000075882 UP000076407 UP000008820 UP000075903 UP000005203 UP000037069 UP000245037 UP000095301 UP000009192 UP000005205 UP000095300 UP000268350 UP000007801 UP000192221 UP000076502 UP000008711 UP000000803 UP000002282
Interpro
CDD
ProteinModelPortal
H9J292
A0A0N0PDE9
A0A2A4K3L9
A0A2W1BKK4
A0A194PVB2
A0A2H1VVD7
+ More
A0A2Z4GWW8 A0A0L7LEL9 A0A2M3ZHQ8 A0A1Q3G591 A0A2M4AJX9 A0A1Q3G587 A0A1L8DEP3 A0A1Q3G595 A0A1I8JU73 A0A1B0GIR5 A0A1I8JT46 A0A1W4X8E4 A0A1C7CZA6 A0A1I8JVP3 A0A1I8JT60 A0A1Y9IHS4 A0A1I8JVP2 A0A1I8JVL0 A0A1W4X876 A0A1W4X6Z7 A0A1W4WXM6 H2L215 A0A023F5C7 Q16P15 A0A1I8JVE1 A0A1I8JVB6 A0A1I8JVB5 A0A0A9VUE7 A0A0K8S3J5 A0A0K8S552 A0A0K8S3X9 A0A146M9C9 A0A0A9W1U4 A0A087ZZW6 V9I9A6 V9IA86 V9I7X5 V9I9S9 A0A0L0C9C4 A0A2P8XHN8 T1PD66 A0A0Q9XIN2 A0A0Q9XKJ7 A0A1B6ES16 W8BPR2 A0A1B6G3Y2 B4KMC3 A0A0Q9XM85 A0A0Q9X8Z8 A0A158NKY1 W8C206 A0A1I8M4N6 A0A1I8M4N7 A0A0Q9XI19 T1PD35 A0A1I8M4N4 A0A1I8QA37 A0A034VI57 A0A1I8QA21 A0A0K8V8W7 A0A0K8UL13 A0A3B0JIV8 A0A1I8Q9X8 A0A034VJ83 A0A3B0IYU5 A0A034VKW0 A0A0K8W0Z0 A0A0P8YAI4 A0A1W4UYZ9 A0A154P501 A0A3B0J2E5 A0A3B0IYK7 A0A034VK61 A0A0P9BX49 B3MCM5 A0A0P8Y2U3 A0A0Q5VP73 A0A1W4UM20 A0A1W4UZA2 A0A1W4UZA7 A0A0J9RJF1 A0A0Q5VQ76 A0A1W4V0K7 V5G4T2 A0A0B4LGD3 A0A0P8XN64 A0A0R1DTU3 A0A1W4ULI7 A0A1W4V0K9 A0A0J9RL32 Q9W1G5 A0A0R1DTT7 A0A0N8P044 A0A0P8XN79
A0A2Z4GWW8 A0A0L7LEL9 A0A2M3ZHQ8 A0A1Q3G591 A0A2M4AJX9 A0A1Q3G587 A0A1L8DEP3 A0A1Q3G595 A0A1I8JU73 A0A1B0GIR5 A0A1I8JT46 A0A1W4X8E4 A0A1C7CZA6 A0A1I8JVP3 A0A1I8JT60 A0A1Y9IHS4 A0A1I8JVP2 A0A1I8JVL0 A0A1W4X876 A0A1W4X6Z7 A0A1W4WXM6 H2L215 A0A023F5C7 Q16P15 A0A1I8JVE1 A0A1I8JVB6 A0A1I8JVB5 A0A0A9VUE7 A0A0K8S3J5 A0A0K8S552 A0A0K8S3X9 A0A146M9C9 A0A0A9W1U4 A0A087ZZW6 V9I9A6 V9IA86 V9I7X5 V9I9S9 A0A0L0C9C4 A0A2P8XHN8 T1PD66 A0A0Q9XIN2 A0A0Q9XKJ7 A0A1B6ES16 W8BPR2 A0A1B6G3Y2 B4KMC3 A0A0Q9XM85 A0A0Q9X8Z8 A0A158NKY1 W8C206 A0A1I8M4N6 A0A1I8M4N7 A0A0Q9XI19 T1PD35 A0A1I8M4N4 A0A1I8QA37 A0A034VI57 A0A1I8QA21 A0A0K8V8W7 A0A0K8UL13 A0A3B0JIV8 A0A1I8Q9X8 A0A034VJ83 A0A3B0IYU5 A0A034VKW0 A0A0K8W0Z0 A0A0P8YAI4 A0A1W4UYZ9 A0A154P501 A0A3B0J2E5 A0A3B0IYK7 A0A034VK61 A0A0P9BX49 B3MCM5 A0A0P8Y2U3 A0A0Q5VP73 A0A1W4UM20 A0A1W4UZA2 A0A1W4UZA7 A0A0J9RJF1 A0A0Q5VQ76 A0A1W4V0K7 V5G4T2 A0A0B4LGD3 A0A0P8XN64 A0A0R1DTU3 A0A1W4ULI7 A0A1W4V0K9 A0A0J9RL32 Q9W1G5 A0A0R1DTT7 A0A0N8P044 A0A0P8XN79
PDB
3GIA
E-value=0.0272416,
Score=89
Ontologies
GO
PANTHER
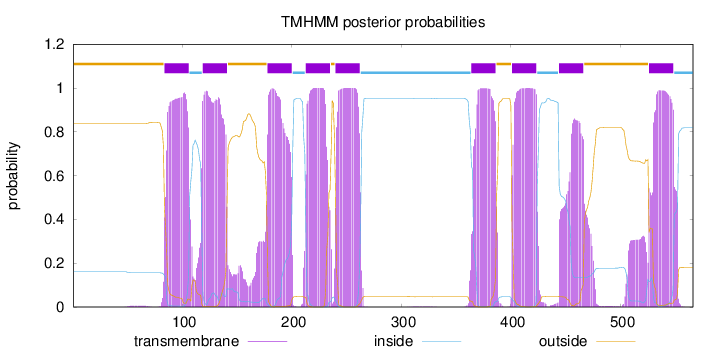
Topology
Length:
566
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
208.54896
Exp number, first 60 AAs:
0.04885
Total prob of N-in:
0.16128
outside
1 - 83
TMhelix
84 - 106
inside
107 - 118
TMhelix
119 - 141
outside
142 - 177
TMhelix
178 - 200
inside
201 - 212
TMhelix
213 - 235
outside
236 - 239
TMhelix
240 - 262
inside
263 - 363
TMhelix
364 - 386
outside
387 - 400
TMhelix
401 - 423
inside
424 - 443
TMhelix
444 - 466
outside
467 - 525
TMhelix
526 - 548
inside
549 - 566
Population Genetic Test Statistics
Pi
292.227814
Theta
191.631145
Tajima's D
1.669647
CLR
0.407001
CSRT
0.820058997050148
Interpretation
Uncertain
