Gene
KWMTBOMO02945
Pre Gene Modal
BGIBMGA003631
Annotation
hypothetical_protein_KGM_15356_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 3.01
Sequence
CDS
ATGCAGTTGAAATGGGGTATAGCAGGCGTGGGAATGATCGCCCATGACTTCCTGACAGCATTAGACACTTTACCTCCGGAACAACACAGGGTTGTTGCAATAGCCGGCAAAGATGTAGATCGCGTTCAACGATTAGCAACACTGCACAAGATCAACACAGCGTACGAAGGTTACGAGCAACTGGCGCGTGATGCTTCTATAGACATTGTTTTCGTGAGTGTTCTGAATTTACAGCACTACGAAATTACAAAGTTAATGCTAGAAAACGATAAACATGTACTCTGTGAGAAGCCAATGGGTATGACTTACAAACAGACTAAATCATTAGTGGACTTGGCGCGTGAGAAGGGTCTGTTCCTTTTAGAAGGAATGTGGTCGAGGTTTTTTCCAGCTTACGACGCATTAGAGAAACATATAACCACTGGCGGCCTTGGTGAAGTCTATCACATCAACGTTCAGTTTGGGGTTCAAATTAACGACATTGAAAGGAATCTGATGAAGGACCTCGGTGGTGGTGCGGTTTTAGACCTAGGAGTGTACATGTTGCAGCTAATAAACTTCATATACAAGGAACCGCCAGTGGATGTAGTGTGCACGGGGCATTTAAGCAAAACGGGAGTAGACGAGTCCGTGTCGTGTGCTTTGAAGTATAAAGACGGAAGAACGGCAACAATGGCCGCCCAGACCCGTGCGACCATGACCAATAAAGCGGAGATTATTGGTACGAAAGGAACAATCTCTTTGGAGTACTTCTGGTGTCCAACGGTATTACACATCTCGGCGGCTAACAGCACCGAATGGACATTACCGAAGGGCAAATACAAGTTCCACTTCCACAACAGCGCTGGACTTAGCTACCAGATACAGGAGTGCTGGGATTGTATTAACAAAGGTATTGTTGAGAGTCCAAAGATGAGCTTAGATGAAAGTATTCTCTTATCCAAATTGACGGACACTATGAGGGCGCAACTAGGAGTACTTGAATCGGAATTATAA
Protein
MQLKWGIAGVGMIAHDFLTALDTLPPEQHRVVAIAGKDVDRVQRLATLHKINTAYEGYEQLARDASIDIVFVSVLNLQHYEITKLMLENDKHVLCEKPMGMTYKQTKSLVDLAREKGLFLLEGMWSRFFPAYDALEKHITTGGLGEVYHINVQFGVQINDIERNLMKDLGGGAVLDLGVYMLQLINFIYKEPPVDVVCTGHLSKTGVDESVSCALKYKDGRTATMAAQTRATMTNKAEIIGTKGTISLEYFWCPTVLHISAANSTEWTLPKGKYKFHFHNSAGLSYQIQECWDCINKGIVESPKMSLDESILLSKLTDTMRAQLGVLESEL
Summary
Uniprot
H9J294
A0A2W1BR00
A0A2A4K3M6
A0A2A4K465
A0A2H1V0D3
S4NR82
+ More
A0A194PQ22 A0A212EX45 A0A0N0PDT1 A0A0L7LFC8 A0A2J7RKP2 A0A067QX71 A0A154PMX1 A0A2A3EAV6 E9I8Q2 H9K9P8 A0A151WJ49 F4W7M8 A0A151IMP4 A0A026W2Y1 A0A158NPM5 A0A0N0BGK4 A0A0C9QG12 A0A195AT36 E2AXQ4 A0A0J7KRK4 A0A0L7R4N4 A0A195EL60 K7J2W9 A0A336MXB9 A0A0K8TTM1 A0A336MS92 A0A195ETJ0 A0A1B0CN82 A0A1W4WJX5 W8BTQ4 B0X5Q2 A0A0K8W9S1 A0A034WTE5 A0A1B0GPG9 A0A1L8E212 A0A0L0BSW4 A0A1L8E2I1 A0A1B0AT96 A0A1A9W5F5 A0A1A9XN38 A0A1I8M199 A0A1I8NNZ7 A0A0T6B2H7 A0A1Q3FQ73 D6W762 A0A1Y1M984 E2BUF6 A0A0A1X461 A0A1A9ZJG4 T1GXU5 A0A1A9UMK4 A0A182PG54 D6W763 A0A1S3KDT8 A0A1B0F9J8 A0A1D2MQR9 A0A1Y1KB79 A0A182PAM0 A0A310SV38 U5EL44 A0A226E6P8 A0A226DXU2 A0A182U3C8 A0A336LLD2 A0A182US82 Q7Q164 Q7PVP0 A0A182HRD8 A0A1S4H0I5 A0A1B6CSQ0 A0A182I5A6 A0A182XKT8 Q16UC6 A0A182UBP2 A0A182GPQ0 A0A182KYZ1 A0A182RG54 A0A1Y1MRP5 A0A084VWF1 A0A182XHT8 A0A182VND1 A0A182QP31 Q29MK9 B4G927 H3AUI9 W5JPK9 A0A1S4FP90 A0A182WHA4 A0A182MC66 A0A182QFW5 A0A194PNI9 A0A2M4BT72 A0A2M4A5K1 B4G928
A0A194PQ22 A0A212EX45 A0A0N0PDT1 A0A0L7LFC8 A0A2J7RKP2 A0A067QX71 A0A154PMX1 A0A2A3EAV6 E9I8Q2 H9K9P8 A0A151WJ49 F4W7M8 A0A151IMP4 A0A026W2Y1 A0A158NPM5 A0A0N0BGK4 A0A0C9QG12 A0A195AT36 E2AXQ4 A0A0J7KRK4 A0A0L7R4N4 A0A195EL60 K7J2W9 A0A336MXB9 A0A0K8TTM1 A0A336MS92 A0A195ETJ0 A0A1B0CN82 A0A1W4WJX5 W8BTQ4 B0X5Q2 A0A0K8W9S1 A0A034WTE5 A0A1B0GPG9 A0A1L8E212 A0A0L0BSW4 A0A1L8E2I1 A0A1B0AT96 A0A1A9W5F5 A0A1A9XN38 A0A1I8M199 A0A1I8NNZ7 A0A0T6B2H7 A0A1Q3FQ73 D6W762 A0A1Y1M984 E2BUF6 A0A0A1X461 A0A1A9ZJG4 T1GXU5 A0A1A9UMK4 A0A182PG54 D6W763 A0A1S3KDT8 A0A1B0F9J8 A0A1D2MQR9 A0A1Y1KB79 A0A182PAM0 A0A310SV38 U5EL44 A0A226E6P8 A0A226DXU2 A0A182U3C8 A0A336LLD2 A0A182US82 Q7Q164 Q7PVP0 A0A182HRD8 A0A1S4H0I5 A0A1B6CSQ0 A0A182I5A6 A0A182XKT8 Q16UC6 A0A182UBP2 A0A182GPQ0 A0A182KYZ1 A0A182RG54 A0A1Y1MRP5 A0A084VWF1 A0A182XHT8 A0A182VND1 A0A182QP31 Q29MK9 B4G927 H3AUI9 W5JPK9 A0A1S4FP90 A0A182WHA4 A0A182MC66 A0A182QFW5 A0A194PNI9 A0A2M4BT72 A0A2M4A5K1 B4G928
Pubmed
19121390
28756777
23622113
26354079
22118469
26227816
+ More
24845553 21282665 21719571 24508170 30249741 21347285 20798317 20075255 26369729 24495485 25348373 26108605 25315136 18362917 19820115 28004739 25830018 27289101 12364791 14747013 17210077 17510324 26483478 20966253 24438588 15632085 17994087 9215903 20920257 23761445
24845553 21282665 21719571 24508170 30249741 21347285 20798317 20075255 26369729 24495485 25348373 26108605 25315136 18362917 19820115 28004739 25830018 27289101 12364791 14747013 17210077 17510324 26483478 20966253 24438588 15632085 17994087 9215903 20920257 23761445
EMBL
BABH01007531
KZ149992
PZC75567.1
NWSH01000167
PCG78857.1
PCG78859.1
+ More
ODYU01000103 SOQ34305.1 GAIX01012961 JAA79599.1 KQ459597 KPI95063.1 AGBW02011836 OWR46059.1 KQ460226 KPJ16469.1 JTDY01001308 KOB74243.1 NEVH01002717 PNF41391.1 KK853253 KDR09314.1 KQ434996 KZC13216.1 KZ288301 PBC28848.1 GL761642 EFZ23042.1 KQ983049 KYQ47851.1 GL887844 EGI69901.1 KQ977026 KYN06220.1 KK107488 QOIP01000012 EZA49946.1 RLU16030.1 ADTU01022615 KQ435775 KOX74942.1 GBYB01002344 JAG72111.1 KQ976745 KYM75383.1 GL443628 EFN61800.1 LBMM01003940 KMQ92973.1 KQ414657 KOC65789.1 KQ978730 KYN28995.1 UFQS01002377 UFQT01002377 SSX13914.1 SSX33333.1 GDAI01000105 JAI17498.1 SSX13913.1 SSX33332.1 KQ981986 KYN31194.1 AJWK01019891 GAMC01003995 GAMC01003992 GAMC01003988 JAC02568.1 DS232392 EDS41033.1 GDHF01028557 GDHF01017237 GDHF01010628 GDHF01009375 GDHF01004408 JAI23757.1 JAI35077.1 JAI41686.1 JAI42939.1 JAI47906.1 GAKP01001350 GAKP01001349 JAC57602.1 AJVK01033660 GFDF01001370 JAV12714.1 JRES01001418 KNC23111.1 GFDF01001369 JAV12715.1 JXJN01003215 JXJN01003216 LJIG01016289 KRT81047.1 GFDL01005284 JAV29761.1 KQ971307 EFA11514.1 GEZM01037072 JAV82213.1 GL450640 EFN80683.1 GBXI01013938 GBXI01008742 GBXI01005282 JAD00354.1 JAD05550.1 JAD09010.1 CAQQ02123182 EFA11515.2 CCAG010019949 LJIJ01000675 ODM95427.1 GEZM01087415 JAV58709.1 KQ759880 OAD62220.1 GANO01001580 JAB58291.1 LNIX01000006 OXA52541.1 LNIX01000009 OXA50093.1 UFQT01000052 SSX19034.1 AAAB01008980 EAA14025.3 AAAB01008984 EAA15029.4 APCN01001694 GEDC01021015 GEDC01004107 JAS16283.1 JAS33191.1 APCN01003842 CH477625 EAT38123.1 JXUM01078870 KQ563093 KXJ74488.1 GEZM01024715 JAV87738.1 ATLV01017583 KE525174 KFB42295.1 AXCN02000825 CH379060 EAL33684.1 CH479180 EDW28857.1 AFYH01076312 AFYH01076313 ADMH02000539 ETN66061.1 AXCM01005520 AXCN02000654 KPI94996.1 GGFJ01007134 MBW56275.1 GGFK01002689 MBW36010.1 EDW28858.1
ODYU01000103 SOQ34305.1 GAIX01012961 JAA79599.1 KQ459597 KPI95063.1 AGBW02011836 OWR46059.1 KQ460226 KPJ16469.1 JTDY01001308 KOB74243.1 NEVH01002717 PNF41391.1 KK853253 KDR09314.1 KQ434996 KZC13216.1 KZ288301 PBC28848.1 GL761642 EFZ23042.1 KQ983049 KYQ47851.1 GL887844 EGI69901.1 KQ977026 KYN06220.1 KK107488 QOIP01000012 EZA49946.1 RLU16030.1 ADTU01022615 KQ435775 KOX74942.1 GBYB01002344 JAG72111.1 KQ976745 KYM75383.1 GL443628 EFN61800.1 LBMM01003940 KMQ92973.1 KQ414657 KOC65789.1 KQ978730 KYN28995.1 UFQS01002377 UFQT01002377 SSX13914.1 SSX33333.1 GDAI01000105 JAI17498.1 SSX13913.1 SSX33332.1 KQ981986 KYN31194.1 AJWK01019891 GAMC01003995 GAMC01003992 GAMC01003988 JAC02568.1 DS232392 EDS41033.1 GDHF01028557 GDHF01017237 GDHF01010628 GDHF01009375 GDHF01004408 JAI23757.1 JAI35077.1 JAI41686.1 JAI42939.1 JAI47906.1 GAKP01001350 GAKP01001349 JAC57602.1 AJVK01033660 GFDF01001370 JAV12714.1 JRES01001418 KNC23111.1 GFDF01001369 JAV12715.1 JXJN01003215 JXJN01003216 LJIG01016289 KRT81047.1 GFDL01005284 JAV29761.1 KQ971307 EFA11514.1 GEZM01037072 JAV82213.1 GL450640 EFN80683.1 GBXI01013938 GBXI01008742 GBXI01005282 JAD00354.1 JAD05550.1 JAD09010.1 CAQQ02123182 EFA11515.2 CCAG010019949 LJIJ01000675 ODM95427.1 GEZM01087415 JAV58709.1 KQ759880 OAD62220.1 GANO01001580 JAB58291.1 LNIX01000006 OXA52541.1 LNIX01000009 OXA50093.1 UFQT01000052 SSX19034.1 AAAB01008980 EAA14025.3 AAAB01008984 EAA15029.4 APCN01001694 GEDC01021015 GEDC01004107 JAS16283.1 JAS33191.1 APCN01003842 CH477625 EAT38123.1 JXUM01078870 KQ563093 KXJ74488.1 GEZM01024715 JAV87738.1 ATLV01017583 KE525174 KFB42295.1 AXCN02000825 CH379060 EAL33684.1 CH479180 EDW28857.1 AFYH01076312 AFYH01076313 ADMH02000539 ETN66061.1 AXCM01005520 AXCN02000654 KPI94996.1 GGFJ01007134 MBW56275.1 GGFK01002689 MBW36010.1 EDW28858.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000235965 UP000027135 UP000076502 UP000242457 UP000005203 UP000075809 UP000007755 UP000078542 UP000053097 UP000279307 UP000005205 UP000053105 UP000078540 UP000000311 UP000036403 UP000053825 UP000078492 UP000002358 UP000078541 UP000092461 UP000192223 UP000002320 UP000092462 UP000037069 UP000092460 UP000091820 UP000092443 UP000095301 UP000095300 UP000007266 UP000008237 UP000092445 UP000015102 UP000078200 UP000075885 UP000085678 UP000092444 UP000094527 UP000198287 UP000075902 UP000075903 UP000007062 UP000075840 UP000076407 UP000008820 UP000069940 UP000249989 UP000075882 UP000075900 UP000030765 UP000075886 UP000001819 UP000008744 UP000008672 UP000000673 UP000075920 UP000075883
UP000235965 UP000027135 UP000076502 UP000242457 UP000005203 UP000075809 UP000007755 UP000078542 UP000053097 UP000279307 UP000005205 UP000053105 UP000078540 UP000000311 UP000036403 UP000053825 UP000078492 UP000002358 UP000078541 UP000092461 UP000192223 UP000002320 UP000092462 UP000037069 UP000092460 UP000091820 UP000092443 UP000095301 UP000095300 UP000007266 UP000008237 UP000092445 UP000015102 UP000078200 UP000075885 UP000085678 UP000092444 UP000094527 UP000198287 UP000075902 UP000075903 UP000007062 UP000075840 UP000076407 UP000008820 UP000069940 UP000249989 UP000075882 UP000075900 UP000030765 UP000075886 UP000001819 UP000008744 UP000008672 UP000000673 UP000075920 UP000075883
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9J294
A0A2W1BR00
A0A2A4K3M6
A0A2A4K465
A0A2H1V0D3
S4NR82
+ More
A0A194PQ22 A0A212EX45 A0A0N0PDT1 A0A0L7LFC8 A0A2J7RKP2 A0A067QX71 A0A154PMX1 A0A2A3EAV6 E9I8Q2 H9K9P8 A0A151WJ49 F4W7M8 A0A151IMP4 A0A026W2Y1 A0A158NPM5 A0A0N0BGK4 A0A0C9QG12 A0A195AT36 E2AXQ4 A0A0J7KRK4 A0A0L7R4N4 A0A195EL60 K7J2W9 A0A336MXB9 A0A0K8TTM1 A0A336MS92 A0A195ETJ0 A0A1B0CN82 A0A1W4WJX5 W8BTQ4 B0X5Q2 A0A0K8W9S1 A0A034WTE5 A0A1B0GPG9 A0A1L8E212 A0A0L0BSW4 A0A1L8E2I1 A0A1B0AT96 A0A1A9W5F5 A0A1A9XN38 A0A1I8M199 A0A1I8NNZ7 A0A0T6B2H7 A0A1Q3FQ73 D6W762 A0A1Y1M984 E2BUF6 A0A0A1X461 A0A1A9ZJG4 T1GXU5 A0A1A9UMK4 A0A182PG54 D6W763 A0A1S3KDT8 A0A1B0F9J8 A0A1D2MQR9 A0A1Y1KB79 A0A182PAM0 A0A310SV38 U5EL44 A0A226E6P8 A0A226DXU2 A0A182U3C8 A0A336LLD2 A0A182US82 Q7Q164 Q7PVP0 A0A182HRD8 A0A1S4H0I5 A0A1B6CSQ0 A0A182I5A6 A0A182XKT8 Q16UC6 A0A182UBP2 A0A182GPQ0 A0A182KYZ1 A0A182RG54 A0A1Y1MRP5 A0A084VWF1 A0A182XHT8 A0A182VND1 A0A182QP31 Q29MK9 B4G927 H3AUI9 W5JPK9 A0A1S4FP90 A0A182WHA4 A0A182MC66 A0A182QFW5 A0A194PNI9 A0A2M4BT72 A0A2M4A5K1 B4G928
A0A194PQ22 A0A212EX45 A0A0N0PDT1 A0A0L7LFC8 A0A2J7RKP2 A0A067QX71 A0A154PMX1 A0A2A3EAV6 E9I8Q2 H9K9P8 A0A151WJ49 F4W7M8 A0A151IMP4 A0A026W2Y1 A0A158NPM5 A0A0N0BGK4 A0A0C9QG12 A0A195AT36 E2AXQ4 A0A0J7KRK4 A0A0L7R4N4 A0A195EL60 K7J2W9 A0A336MXB9 A0A0K8TTM1 A0A336MS92 A0A195ETJ0 A0A1B0CN82 A0A1W4WJX5 W8BTQ4 B0X5Q2 A0A0K8W9S1 A0A034WTE5 A0A1B0GPG9 A0A1L8E212 A0A0L0BSW4 A0A1L8E2I1 A0A1B0AT96 A0A1A9W5F5 A0A1A9XN38 A0A1I8M199 A0A1I8NNZ7 A0A0T6B2H7 A0A1Q3FQ73 D6W762 A0A1Y1M984 E2BUF6 A0A0A1X461 A0A1A9ZJG4 T1GXU5 A0A1A9UMK4 A0A182PG54 D6W763 A0A1S3KDT8 A0A1B0F9J8 A0A1D2MQR9 A0A1Y1KB79 A0A182PAM0 A0A310SV38 U5EL44 A0A226E6P8 A0A226DXU2 A0A182U3C8 A0A336LLD2 A0A182US82 Q7Q164 Q7PVP0 A0A182HRD8 A0A1S4H0I5 A0A1B6CSQ0 A0A182I5A6 A0A182XKT8 Q16UC6 A0A182UBP2 A0A182GPQ0 A0A182KYZ1 A0A182RG54 A0A1Y1MRP5 A0A084VWF1 A0A182XHT8 A0A182VND1 A0A182QP31 Q29MK9 B4G927 H3AUI9 W5JPK9 A0A1S4FP90 A0A182WHA4 A0A182MC66 A0A182QFW5 A0A194PNI9 A0A2M4BT72 A0A2M4A5K1 B4G928
PDB
3OHS
E-value=3.50188e-55,
Score=543
Ontologies
PATHWAY
GO
Topology
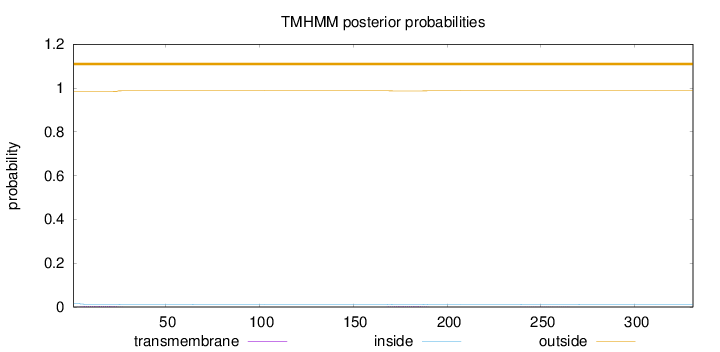
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13489
Exp number, first 60 AAs:
0.0759
Total prob of N-in:
0.01604
outside
1 - 331
Population Genetic Test Statistics
Pi
210.462429
Theta
182.754911
Tajima's D
0.488021
CLR
0.11542
CSRT
0.511724413779311
Interpretation
Uncertain
