Gene
KWMTBOMO02944 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003564
Annotation
PREDICTED:_ornithine_aminotransferase?_mitochondrial_[Papilio_polytes]
Full name
Ornithine aminotransferase
+ More
Ornithine aminotransferase, mitochondrial
Ornithine aminotransferase, mitochondrial
Alternative Name
Ornithine--oxo-acid aminotransferase
Location in the cell
Cytoplasmic Reliability : 2.729
Sequence
CDS
ATGGCTGAACAAAATTTGTCATCTAAAGAAATTTTCCAGTTGGAAGACAAATATGGTTGTAGAAATTACGCTCCCTTGCCTGTTGCTTTATGCCGAGGTGAAGGAGTGTTCGTGTGGGATGTCGAAGGAAAAAAGTATTATGATTTTTTAAGCGCCTACTCCGCAGTTAACCAAGGTCACTGCCACCCTAGAATAATTGAAGCTTTGAAGAAACAAGCTGATAATTTGACACTAGTGTCAAGAGCATTTTATTCCGATCAACTTGGCAAGTATGAAAAGTATATGACAGAGTTATTTGGATATGATCGCCTGCTTCCTATGAACACGGGTGTTGAAGGAGGTGAAAGTGCATGCAAGATAGCACGTAAATGGGGGTACGAAGTCAAGAAGATACCTGAAGGCCAAGCTAAGATAATTTTTGCGGAAGGAAACTTCTGGGGGCGAACTCTATCGGCGGTATCATCATCATCAGACCCAACCTGCTACCAGGGCTTCGGACCGTACATGCCGGGCTTCATTCTTATTCCCTACAACAACATCCCAGCTTTAGAGAAAGCTCTCCAAGATCCGACAGTCGCGGCTTACATGGTTGAACCGATTCAAGGAGAGGCAGGCGTCGTAATACCTGACGACGGCTATCTCAAGAAGGTTCGAGAGCTATGTACAAAGCACAACGTGTTGTGGATTGCGGACGAAGTACAGACTGGATTAGGTCGCACAGGCAAGTTGTTGGCTGTGGAACACGAAGGCGTGAAACCAGACATACTAATACTTGGCAAAGCACTCAGCGGGGGTGTCTTGCCTGTTTCCGCTGTTCTAACTAGCAACGACATTATGGATGTGATAAAGCCAGGCACTCACGGATCAACGTACGGAGGAAACCCTTTGGCGTGCGCTGTAGCCACAGAAGCTATTAAGGTGCTACTGGATGAGAAATTATCGGAAAACGCTGAGAGGATGGGCAAAATACTACGCGAAGAGCTGAGTCAGATACCAAAAACACAGATTCGTACAGTTCGAGGGAGGGGACTGATGTGTGCTATTGTAGTGGATGACAGCATTCCAGCATCGGAGTTGTGTCTCCGCCTGCGCGACAATGGCCTGCTAGCGAAGCCGACGCACGGACAGACCGTGCGCCTCGCGCCGCCGCTCGTCATCACAGAGGCACAGATACGAGACGGCGCTGATATCATCAGGAACGTCTTCCAAAGCTTTCAAAAATAA
Protein
MAEQNLSSKEIFQLEDKYGCRNYAPLPVALCRGEGVFVWDVEGKKYYDFLSAYSAVNQGHCHPRIIEALKKQADNLTLVSRAFYSDQLGKYEKYMTELFGYDRLLPMNTGVEGGESACKIARKWGYEVKKIPEGQAKIIFAEGNFWGRTLSAVSSSSDPTCYQGFGPYMPGFILIPYNNIPALEKALQDPTVAAYMVEPIQGEAGVVIPDDGYLKKVRELCTKHNVLWIADEVQTGLGRTGKLLAVEHEGVKPDILILGKALSGGVLPVSAVLTSNDIMDVIKPGTHGSTYGGNPLACAVATEAIKVLLDEKLSENAERMGKILREELSQIPKTQIRTVRGRGLMCAIVVDDSIPASELCLRLRDNGLLAKPTHGQTVRLAPPLVITEAQIRDGADIIRNVFQSFQK
Summary
Catalytic Activity
a 2-oxocarboxylate + L-ornithine = an L-alpha-amino acid + L-glutamate 5-semialdehyde
Cofactor
pyridoxal 5'-phosphate
Subunit
Homotetramer.
Similarity
Belongs to the class-III pyridoxal-phosphate-dependent aminotransferase family.
Keywords
Aminotransferase
Complete proteome
Mitochondrion
Pyridoxal phosphate
Reference proteome
Transferase
Transit peptide
Feature
chain Ornithine aminotransferase, mitochondrial
Uniprot
H9J227
A0A2W1BSE7
A0A2H1V208
A0A2A4K599
A0A194PP28
A0A2A4K504
+ More
A0A1B6EZ27 A0A1B6GNX8 A0A068TKS2 A0A026W5N0 A0A3P8U0N8 A0A023ESS8 E9FVF0 A0A0S7KPC2 A0A067QWR3 A0A0F8AV93 A0A3B4AQM1 A0A2J7QY62 A0A3B4AS56 A0A1Q3F533 A0A2J7QY42 Q17AI0 A0A1Q3F622 A0A1Q3F698 A0A182GND5 E6ZHM0 A0A3P9NL21 A0A146X528 A0A2I4B428 A0A3P9NL53 A0A087XZD9 A0A1S4FA38 A0A1Q3F3S2 A0A3B3WLE1 A0A182NNF6 A0A1Q3F4V9 A0A1A8KFG0 A0A1A8BFP7 A0A1A7Z8J7 A0A182HXL2 A0A1A8RPL5 A0A1A8PMT2 A0A182YI01 A0A182FLV5 Q7Q2S8 A0A182WYI0 A0A182V1W3 A0A1A7Z2N1 B0WBA3 A0A182QX32 A0A3B4YQR4 A0A182WCP7 A7UUY2 A0A1A8G267 A0A182RKN2 A0A3B4V522 G3NDH0 A0A182MDK4 A0A0N0BK08 A0A182LI44 A0A034WFQ4 M4AQ51 A0A182SLB7 A0A2C9JJQ0 T1PDL2 A0A210PIA6 A0A1B6ML76 A0A1B6MT38 A0A1I8NTV1 A0A336K6D8 A0A182J635 A0A146X2X9 A0A3B3BXQ4 A0A1I8NBJ2 A0A0K8VPG6 A0A3B5B483 A0A2U9CTF9 A0A1D2M6T4 A0A3P9K9K3 H2M0Y8 A0A068XGC8 K1Q1I3 A0A3B1J0N7 A0A3P9IXL5 A0A1S3QJA5 B5X1S8 A0A3P9IXN2 A0A2D0S7M8 A0A1S3N5V9 A0A0L0C7X4 A0A3P8WPY9 Q7ZX40 A0A0P4VUE2 F4WR60 A0A310S4E6 B4IIP0 Q28FA4 A0A1L8FIZ3 Q9VW26 F8W5L3
A0A1B6EZ27 A0A1B6GNX8 A0A068TKS2 A0A026W5N0 A0A3P8U0N8 A0A023ESS8 E9FVF0 A0A0S7KPC2 A0A067QWR3 A0A0F8AV93 A0A3B4AQM1 A0A2J7QY62 A0A3B4AS56 A0A1Q3F533 A0A2J7QY42 Q17AI0 A0A1Q3F622 A0A1Q3F698 A0A182GND5 E6ZHM0 A0A3P9NL21 A0A146X528 A0A2I4B428 A0A3P9NL53 A0A087XZD9 A0A1S4FA38 A0A1Q3F3S2 A0A3B3WLE1 A0A182NNF6 A0A1Q3F4V9 A0A1A8KFG0 A0A1A8BFP7 A0A1A7Z8J7 A0A182HXL2 A0A1A8RPL5 A0A1A8PMT2 A0A182YI01 A0A182FLV5 Q7Q2S8 A0A182WYI0 A0A182V1W3 A0A1A7Z2N1 B0WBA3 A0A182QX32 A0A3B4YQR4 A0A182WCP7 A7UUY2 A0A1A8G267 A0A182RKN2 A0A3B4V522 G3NDH0 A0A182MDK4 A0A0N0BK08 A0A182LI44 A0A034WFQ4 M4AQ51 A0A182SLB7 A0A2C9JJQ0 T1PDL2 A0A210PIA6 A0A1B6ML76 A0A1B6MT38 A0A1I8NTV1 A0A336K6D8 A0A182J635 A0A146X2X9 A0A3B3BXQ4 A0A1I8NBJ2 A0A0K8VPG6 A0A3B5B483 A0A2U9CTF9 A0A1D2M6T4 A0A3P9K9K3 H2M0Y8 A0A068XGC8 K1Q1I3 A0A3B1J0N7 A0A3P9IXL5 A0A1S3QJA5 B5X1S8 A0A3P9IXN2 A0A2D0S7M8 A0A1S3N5V9 A0A0L0C7X4 A0A3P8WPY9 Q7ZX40 A0A0P4VUE2 F4WR60 A0A310S4E6 B4IIP0 Q28FA4 A0A1L8FIZ3 Q9VW26 F8W5L3
EC Number
2.6.1.13
Pubmed
19121390
28756777
26354079
24508170
24945155
21292972
+ More
24845553 25835551 25463417 17510324 26483478 25244985 12364791 14747013 17210077 20966253 25348373 23542700 15562597 28812685 29451363 25315136 27289101 17554307 22992520 25329095 20433749 26108605 24487278 21719571 17994087 20431018 27762356 10731132 12537572 12537569 23594743
24845553 25835551 25463417 17510324 26483478 25244985 12364791 14747013 17210077 20966253 25348373 23542700 15562597 28812685 29451363 25315136 27289101 17554307 22992520 25329095 20433749 26108605 24487278 21719571 17994087 20431018 27762356 10731132 12537572 12537569 23594743
EMBL
BABH01007531
BABH01007532
KZ149992
PZC75566.1
ODYU01000103
SOQ34304.1
+ More
NWSH01000167 PCG78852.1 KQ459597 KPI95062.1 PCG78853.1 GECZ01026564 JAS43205.1 GECZ01005650 JAS64119.1 HG965792 CDO39388.1 KK107390 EZA51382.1 GAPW01001255 JAC12343.1 GL732525 EFX88542.1 GBYX01202367 JAO79073.1 KK852923 KDR13737.1 KQ042638 KKF12738.1 NEVH01009368 PNF33504.1 GFDL01012406 JAV22639.1 PNF33503.1 CH477335 EAT43242.1 GFDL01012030 JAV23015.1 GFDL01012012 JAV23033.1 JXUM01076259 KQ562935 KXJ74780.1 FQ310507 CBN81554.1 GCES01049881 JAR36442.1 AYCK01001153 GFDL01012897 JAV22148.1 GFDL01012445 JAV22600.1 HAED01000950 HAEE01010973 SBR31023.1 HADZ01001787 HAEA01002762 SBP65728.1 HADY01000333 HAEJ01003459 SBP38818.1 APCN01005321 HAEH01005690 HAEI01009507 SBS07971.1 HAEF01007062 HAEG01008612 SBR82314.1 AAAB01008968 EAA13194.4 HADW01001343 HADX01014875 SBP37107.1 DS231878 EDS42247.1 AXCN02001909 EDO63496.1 HAEB01018780 HAEC01000977 SBQ65307.1 AXCM01006583 KQ435710 KOX79798.1 GAKP01005425 GAKP01005424 GAKP01005423 GAKP01005422 JAC53529.1 KA646784 AFP61413.1 NEDP02076661 OWF36207.1 GEBQ01003352 JAT36625.1 GEBQ01000928 JAT39049.1 UFQS01000118 UFQT01000118 SSX00031.1 SSX20411.1 GCES01049882 JAR36441.1 GDHF01011540 JAI40774.1 CP026260 AWP17882.1 LJIJ01003321 ODM88683.1 JH815769 EKC25219.1 BT044997 ACI33259.1 JRES01000760 KNC28518.1 BC045249 AAH45249.1 GDRN01096103 JAI59403.1 GL888284 EGI63287.1 KQ777939 OAD52025.1 CH480844 EDW49811.1 AAMC01073582 BC167123 BC167345 CR762076 AAI67123.1 AAI67345.1 CAJ81924.1 CM004478 OCT71560.1 AE014296 AY047517 AL953868 CABZ01057903 CABZ01057904
NWSH01000167 PCG78852.1 KQ459597 KPI95062.1 PCG78853.1 GECZ01026564 JAS43205.1 GECZ01005650 JAS64119.1 HG965792 CDO39388.1 KK107390 EZA51382.1 GAPW01001255 JAC12343.1 GL732525 EFX88542.1 GBYX01202367 JAO79073.1 KK852923 KDR13737.1 KQ042638 KKF12738.1 NEVH01009368 PNF33504.1 GFDL01012406 JAV22639.1 PNF33503.1 CH477335 EAT43242.1 GFDL01012030 JAV23015.1 GFDL01012012 JAV23033.1 JXUM01076259 KQ562935 KXJ74780.1 FQ310507 CBN81554.1 GCES01049881 JAR36442.1 AYCK01001153 GFDL01012897 JAV22148.1 GFDL01012445 JAV22600.1 HAED01000950 HAEE01010973 SBR31023.1 HADZ01001787 HAEA01002762 SBP65728.1 HADY01000333 HAEJ01003459 SBP38818.1 APCN01005321 HAEH01005690 HAEI01009507 SBS07971.1 HAEF01007062 HAEG01008612 SBR82314.1 AAAB01008968 EAA13194.4 HADW01001343 HADX01014875 SBP37107.1 DS231878 EDS42247.1 AXCN02001909 EDO63496.1 HAEB01018780 HAEC01000977 SBQ65307.1 AXCM01006583 KQ435710 KOX79798.1 GAKP01005425 GAKP01005424 GAKP01005423 GAKP01005422 JAC53529.1 KA646784 AFP61413.1 NEDP02076661 OWF36207.1 GEBQ01003352 JAT36625.1 GEBQ01000928 JAT39049.1 UFQS01000118 UFQT01000118 SSX00031.1 SSX20411.1 GCES01049882 JAR36441.1 GDHF01011540 JAI40774.1 CP026260 AWP17882.1 LJIJ01003321 ODM88683.1 JH815769 EKC25219.1 BT044997 ACI33259.1 JRES01000760 KNC28518.1 BC045249 AAH45249.1 GDRN01096103 JAI59403.1 GL888284 EGI63287.1 KQ777939 OAD52025.1 CH480844 EDW49811.1 AAMC01073582 BC167123 BC167345 CR762076 AAI67123.1 AAI67345.1 CAJ81924.1 CM004478 OCT71560.1 AE014296 AY047517 AL953868 CABZ01057903 CABZ01057904
Proteomes
UP000005204
UP000218220
UP000053268
UP000053097
UP000265080
UP000000305
+ More
UP000027135 UP000261520 UP000235965 UP000008820 UP000069940 UP000249989 UP000242638 UP000192220 UP000028760 UP000261480 UP000075884 UP000075840 UP000076408 UP000069272 UP000007062 UP000076407 UP000075903 UP000002320 UP000075886 UP000261360 UP000075920 UP000075900 UP000261420 UP000007635 UP000075883 UP000053105 UP000075882 UP000002852 UP000075901 UP000076420 UP000242188 UP000095300 UP000075880 UP000261560 UP000095301 UP000261400 UP000246464 UP000094527 UP000265180 UP000001038 UP000005408 UP000018467 UP000265200 UP000087266 UP000221080 UP000037069 UP000265120 UP000007755 UP000001292 UP000008143 UP000186698 UP000000803 UP000000437
UP000027135 UP000261520 UP000235965 UP000008820 UP000069940 UP000249989 UP000242638 UP000192220 UP000028760 UP000261480 UP000075884 UP000075840 UP000076408 UP000069272 UP000007062 UP000076407 UP000075903 UP000002320 UP000075886 UP000261360 UP000075920 UP000075900 UP000261420 UP000007635 UP000075883 UP000053105 UP000075882 UP000002852 UP000075901 UP000076420 UP000242188 UP000095300 UP000075880 UP000261560 UP000095301 UP000261400 UP000246464 UP000094527 UP000265180 UP000001038 UP000005408 UP000018467 UP000265200 UP000087266 UP000221080 UP000037069 UP000265120 UP000007755 UP000001292 UP000008143 UP000186698 UP000000803 UP000000437
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
H9J227
A0A2W1BSE7
A0A2H1V208
A0A2A4K599
A0A194PP28
A0A2A4K504
+ More
A0A1B6EZ27 A0A1B6GNX8 A0A068TKS2 A0A026W5N0 A0A3P8U0N8 A0A023ESS8 E9FVF0 A0A0S7KPC2 A0A067QWR3 A0A0F8AV93 A0A3B4AQM1 A0A2J7QY62 A0A3B4AS56 A0A1Q3F533 A0A2J7QY42 Q17AI0 A0A1Q3F622 A0A1Q3F698 A0A182GND5 E6ZHM0 A0A3P9NL21 A0A146X528 A0A2I4B428 A0A3P9NL53 A0A087XZD9 A0A1S4FA38 A0A1Q3F3S2 A0A3B3WLE1 A0A182NNF6 A0A1Q3F4V9 A0A1A8KFG0 A0A1A8BFP7 A0A1A7Z8J7 A0A182HXL2 A0A1A8RPL5 A0A1A8PMT2 A0A182YI01 A0A182FLV5 Q7Q2S8 A0A182WYI0 A0A182V1W3 A0A1A7Z2N1 B0WBA3 A0A182QX32 A0A3B4YQR4 A0A182WCP7 A7UUY2 A0A1A8G267 A0A182RKN2 A0A3B4V522 G3NDH0 A0A182MDK4 A0A0N0BK08 A0A182LI44 A0A034WFQ4 M4AQ51 A0A182SLB7 A0A2C9JJQ0 T1PDL2 A0A210PIA6 A0A1B6ML76 A0A1B6MT38 A0A1I8NTV1 A0A336K6D8 A0A182J635 A0A146X2X9 A0A3B3BXQ4 A0A1I8NBJ2 A0A0K8VPG6 A0A3B5B483 A0A2U9CTF9 A0A1D2M6T4 A0A3P9K9K3 H2M0Y8 A0A068XGC8 K1Q1I3 A0A3B1J0N7 A0A3P9IXL5 A0A1S3QJA5 B5X1S8 A0A3P9IXN2 A0A2D0S7M8 A0A1S3N5V9 A0A0L0C7X4 A0A3P8WPY9 Q7ZX40 A0A0P4VUE2 F4WR60 A0A310S4E6 B4IIP0 Q28FA4 A0A1L8FIZ3 Q9VW26 F8W5L3
A0A1B6EZ27 A0A1B6GNX8 A0A068TKS2 A0A026W5N0 A0A3P8U0N8 A0A023ESS8 E9FVF0 A0A0S7KPC2 A0A067QWR3 A0A0F8AV93 A0A3B4AQM1 A0A2J7QY62 A0A3B4AS56 A0A1Q3F533 A0A2J7QY42 Q17AI0 A0A1Q3F622 A0A1Q3F698 A0A182GND5 E6ZHM0 A0A3P9NL21 A0A146X528 A0A2I4B428 A0A3P9NL53 A0A087XZD9 A0A1S4FA38 A0A1Q3F3S2 A0A3B3WLE1 A0A182NNF6 A0A1Q3F4V9 A0A1A8KFG0 A0A1A8BFP7 A0A1A7Z8J7 A0A182HXL2 A0A1A8RPL5 A0A1A8PMT2 A0A182YI01 A0A182FLV5 Q7Q2S8 A0A182WYI0 A0A182V1W3 A0A1A7Z2N1 B0WBA3 A0A182QX32 A0A3B4YQR4 A0A182WCP7 A7UUY2 A0A1A8G267 A0A182RKN2 A0A3B4V522 G3NDH0 A0A182MDK4 A0A0N0BK08 A0A182LI44 A0A034WFQ4 M4AQ51 A0A182SLB7 A0A2C9JJQ0 T1PDL2 A0A210PIA6 A0A1B6ML76 A0A1B6MT38 A0A1I8NTV1 A0A336K6D8 A0A182J635 A0A146X2X9 A0A3B3BXQ4 A0A1I8NBJ2 A0A0K8VPG6 A0A3B5B483 A0A2U9CTF9 A0A1D2M6T4 A0A3P9K9K3 H2M0Y8 A0A068XGC8 K1Q1I3 A0A3B1J0N7 A0A3P9IXL5 A0A1S3QJA5 B5X1S8 A0A3P9IXN2 A0A2D0S7M8 A0A1S3N5V9 A0A0L0C7X4 A0A3P8WPY9 Q7ZX40 A0A0P4VUE2 F4WR60 A0A310S4E6 B4IIP0 Q28FA4 A0A1L8FIZ3 Q9VW26 F8W5L3
PDB
2OAT
E-value=1.19387e-163,
Score=1479
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion matrix
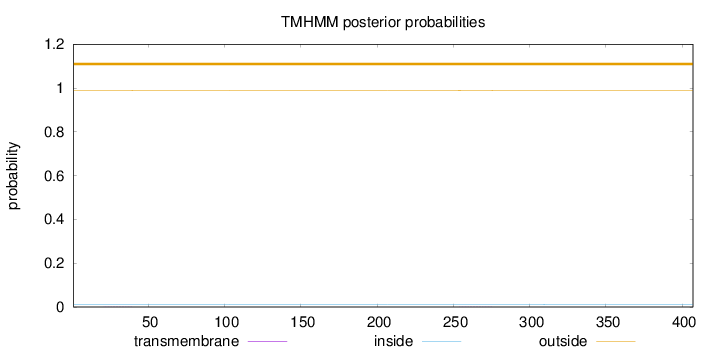
Length:
407
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02066
Exp number, first 60 AAs:
0.00351
Total prob of N-in:
0.01045
outside
1 - 407
Population Genetic Test Statistics
Pi
189.040199
Theta
176.570262
Tajima's D
0.430017
CLR
1.073894
CSRT
0.494525273736313
Interpretation
Uncertain
