Gene
KWMTBOMO02943
Pre Gene Modal
BGIBMGA003563
Annotation
PREDICTED:_mimitin?_mitochondrial_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.151 Mitochondrial Reliability : 1.391 Nuclear Reliability : 1.27
Sequence
CDS
ATGGCAGTTTTTGTCTGTACTATGTCTAATGGAGAATATCGACAAGTATGGCGCATTGTTTTTAGAAATTTCATTAATTCACTTAGACCAAAACAGGTTACTGGAAACAATGTTGGTAAAGATTACATAGGTAACACTTATTTTGAAATACCTGCTAGTCCAAGTGAAGGTAAAAGAAAGCCATCACGTTGGTATGATCCACCTAAAGGTCAGGATTTCCAAAATCCAATACCAGCCGAATGGGAGTCCTGGTTGAGAATGAGAAGAAAAGAACCACCATCAGAAGAGGAAATAGCAAAAAATGTTGCTATAGCCCAAATAAAGAAGGAGAATGCTGCTAAAATTGAAATGAAGAGGCTTGCAGAGGGCGGATCTCTCCCGGCAGTTCCAGAGAGAGGGCCTCAGTCGTACCCAACTTATGATGAATATTCTACTGGAGACTCAGAAGGTGTCCACTCAAACAAAAAATGA
Protein
MAVFVCTMSNGEYRQVWRIVFRNFINSLRPKQVTGNNVGKDYIGNTYFEIPASPSEGKRKPSRWYDPPKGQDFQNPIPAEWESWLRMRRKEPPSEEEIAKNVAIAQIKKENAAKIEMKRLAEGGSLPAVPERGPQSYPTYDEYSTGDSEGVHSNKK
Summary
Uniprot
H9J226
A0A1E1WKG1
A0A1E1WGE6
A0A2W1BPS5
A0A2A4K456
A0A212FEZ7
+ More
A0A2H1V1S6 A0A0N1IB01 A0A194PNQ7 V5G9Y9 D6WAK3 A0A182MYC3 A0A067R041 A0A182GYX2 A0A023EFC6 A0A0K8TQA0 A0A182PTJ9 A0A182QM87 A0A1S4F7K0 A0A182WUS0 A0NDZ8 A0A182HFT0 A0A2J7PC16 Q17CW4 W5JI25 T1PI10 A0A182VTA5 A0A182JFP3 A0A182MC23 A0A084VKV8 A0A1A9ZDD4 A0A1B0F9P0 A0A2M3ZJM3 A0A0P6J945 A0A2M4C2W0 A0A2M4C2F7 A0A182YA21 A0A182SAY7 A0A182FUJ0 A0A1W4UW53 A0A0L0C5X9 A0A0J9R6D0 B3NKT6 M9NEW0 A0A2M4AWA9 A0A182JXL6 A0A1A9V9S9 A0A226F1Y1 A0A0P5R385 B4P6Y7 A0A1Q3FLC0 A0A1B0B7P6 A0A1A9XL35 B4IFL6 A0A0N8DNU6 A0A1D2MPN6 A0A0P9A3Q8 A0A1Y1N7Z6 A0A1Q3FLI5 A0A0P5ZP51 A0A1A9WKT7 A0A182RLQ9 A0A034WHS5 A0A0C9RLB6 A0A1I8QBF6 A0A2R5LHK4 A0A3B0KEX3 A0A1L8EBA6 A0A1L8EC29 A0A1J1IVS6 A0A0K8U1R0 B4GJJ4 B5DHS6 J3JU75 A0A1D2M1V2 W8C876 B4JDW2 B4N7C7 A0A1W4X6C5 A0A1B0CG69 B0X4Y7 A0A1S3D3I4 A0A0L7KSC4 A0A0M3QU68 A0A131Y8F5 A0A293MS95 V5I4X2 A0A0K8R9J7 B4KF48 A0A1L8E161 A0A1D2AJ99 B7PMP8 A0A131XS76 A0A224XNC1 A0A224YUI2 A0A069DNV6 G3MQ45
A0A2H1V1S6 A0A0N1IB01 A0A194PNQ7 V5G9Y9 D6WAK3 A0A182MYC3 A0A067R041 A0A182GYX2 A0A023EFC6 A0A0K8TQA0 A0A182PTJ9 A0A182QM87 A0A1S4F7K0 A0A182WUS0 A0NDZ8 A0A182HFT0 A0A2J7PC16 Q17CW4 W5JI25 T1PI10 A0A182VTA5 A0A182JFP3 A0A182MC23 A0A084VKV8 A0A1A9ZDD4 A0A1B0F9P0 A0A2M3ZJM3 A0A0P6J945 A0A2M4C2W0 A0A2M4C2F7 A0A182YA21 A0A182SAY7 A0A182FUJ0 A0A1W4UW53 A0A0L0C5X9 A0A0J9R6D0 B3NKT6 M9NEW0 A0A2M4AWA9 A0A182JXL6 A0A1A9V9S9 A0A226F1Y1 A0A0P5R385 B4P6Y7 A0A1Q3FLC0 A0A1B0B7P6 A0A1A9XL35 B4IFL6 A0A0N8DNU6 A0A1D2MPN6 A0A0P9A3Q8 A0A1Y1N7Z6 A0A1Q3FLI5 A0A0P5ZP51 A0A1A9WKT7 A0A182RLQ9 A0A034WHS5 A0A0C9RLB6 A0A1I8QBF6 A0A2R5LHK4 A0A3B0KEX3 A0A1L8EBA6 A0A1L8EC29 A0A1J1IVS6 A0A0K8U1R0 B4GJJ4 B5DHS6 J3JU75 A0A1D2M1V2 W8C876 B4JDW2 B4N7C7 A0A1W4X6C5 A0A1B0CG69 B0X4Y7 A0A1S3D3I4 A0A0L7KSC4 A0A0M3QU68 A0A131Y8F5 A0A293MS95 V5I4X2 A0A0K8R9J7 B4KF48 A0A1L8E161 A0A1D2AJ99 B7PMP8 A0A131XS76 A0A224XNC1 A0A224YUI2 A0A069DNV6 G3MQ45
Pubmed
19121390
28756777
22118469
26354079
18362917
19820115
+ More
24845553 26483478 24945155 26369729 12364791 14747013 17210077 17510324 20920257 23761445 25315136 24438588 25244985 26108605 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27289101 28004739 25348373 15632085 22516182 23537049 24495485 26227816 25765539 28049606 28797301 26334808 22216098
24845553 26483478 24945155 26369729 12364791 14747013 17210077 17510324 20920257 23761445 25315136 24438588 25244985 26108605 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27289101 28004739 25348373 15632085 22516182 23537049 24495485 26227816 25765539 28049606 28797301 26334808 22216098
EMBL
BABH01007533
GDQN01003589
JAT87465.1
GDQN01004981
GDQN01002446
GDQN01001595
+ More
JAT86073.1 JAT88608.1 JAT89459.1 KZ149992 PZC75565.1 NWSH01000167 PCG78849.1 AGBW02008872 OWR52319.1 ODYU01000103 SOQ34302.1 KQ460226 KPJ16467.1 KQ459597 KPI95061.1 GALX01007637 JAB60829.1 KQ971312 EEZ98642.1 KK853137 KDR10793.1 JXUM01098411 KQ564418 KXJ72316.1 GAPW01005586 JAC08012.1 GDAI01001041 JAI16562.1 AXCN02002050 AAAB01008933 EAU76757.1 APCN01005572 NEVH01027073 PNF13872.1 CH477303 EAT44210.1 ADMH02001479 ETN62419.1 KA647770 AFP62399.1 AXCM01000338 ATLV01014308 KE524956 KFB38602.1 CCAG010005373 GGFM01007951 MBW28702.1 GDIQ01021205 JAN73532.1 GGFJ01010350 MBW59491.1 GGFJ01010351 MBW59492.1 JRES01000842 KNC27803.1 CM002910 KMY91294.1 CH954179 EDV54459.1 AE014134 AY051903 AFH03795.1 AGW52171.1 GGFK01011671 MBW44992.1 LNIX01000001 OXA63478.1 GDIQ01126105 JAL25621.1 CM000158 EDW89956.1 GFDL01006720 JAV28325.1 JXJN01009697 CH480833 EDW46438.1 GDIP01015479 JAM88236.1 LJIJ01000761 ODM94735.1 CH902620 KPU73191.1 GEZM01010526 GEZM01010525 JAV94011.1 GFDL01006667 JAV28378.1 GDIP01054326 JAM49389.1 GAKP01005272 JAC53680.1 GBYB01007711 JAG77478.1 GGLE01004875 MBY09001.1 OUUW01000006 SPP82158.1 GFDG01002904 JAV15895.1 GFDG01002627 JAV16172.1 CVRI01000062 CRL04208.1 GDHF01032074 JAI20240.1 CH479184 EDW36810.1 CH379058 EDY69853.1 BT126788 KB631815 AEE61750.1 ERL86438.1 LJIJ01006756 ODM86922.1 GAMC01000092 JAC06464.1 CH916368 EDW03482.1 CH964182 EDW80268.1 AJWK01010790 AJWK01010791 AJWK01010792 DS232359 EDS40602.1 JTDY01006277 KOB66148.1 CP012523 ALC40109.1 GEFM01000699 JAP75097.1 GFWV01018976 MAA43704.1 GANP01000875 JAB83593.1 GADI01005966 JAA67842.1 CH933807 EDW13031.1 GFDF01001647 JAV12437.1 GETE01000079 JAT79266.1 ABJB010110070 ABJB010413306 ABJB010704850 DS748754 EEC07870.1 GEFH01000055 JAP68526.1 GFTR01002439 JAW13987.1 GFPF01010121 MAA21267.1 GBGD01003379 JAC85510.1 JO843996 AEO35613.1
JAT86073.1 JAT88608.1 JAT89459.1 KZ149992 PZC75565.1 NWSH01000167 PCG78849.1 AGBW02008872 OWR52319.1 ODYU01000103 SOQ34302.1 KQ460226 KPJ16467.1 KQ459597 KPI95061.1 GALX01007637 JAB60829.1 KQ971312 EEZ98642.1 KK853137 KDR10793.1 JXUM01098411 KQ564418 KXJ72316.1 GAPW01005586 JAC08012.1 GDAI01001041 JAI16562.1 AXCN02002050 AAAB01008933 EAU76757.1 APCN01005572 NEVH01027073 PNF13872.1 CH477303 EAT44210.1 ADMH02001479 ETN62419.1 KA647770 AFP62399.1 AXCM01000338 ATLV01014308 KE524956 KFB38602.1 CCAG010005373 GGFM01007951 MBW28702.1 GDIQ01021205 JAN73532.1 GGFJ01010350 MBW59491.1 GGFJ01010351 MBW59492.1 JRES01000842 KNC27803.1 CM002910 KMY91294.1 CH954179 EDV54459.1 AE014134 AY051903 AFH03795.1 AGW52171.1 GGFK01011671 MBW44992.1 LNIX01000001 OXA63478.1 GDIQ01126105 JAL25621.1 CM000158 EDW89956.1 GFDL01006720 JAV28325.1 JXJN01009697 CH480833 EDW46438.1 GDIP01015479 JAM88236.1 LJIJ01000761 ODM94735.1 CH902620 KPU73191.1 GEZM01010526 GEZM01010525 JAV94011.1 GFDL01006667 JAV28378.1 GDIP01054326 JAM49389.1 GAKP01005272 JAC53680.1 GBYB01007711 JAG77478.1 GGLE01004875 MBY09001.1 OUUW01000006 SPP82158.1 GFDG01002904 JAV15895.1 GFDG01002627 JAV16172.1 CVRI01000062 CRL04208.1 GDHF01032074 JAI20240.1 CH479184 EDW36810.1 CH379058 EDY69853.1 BT126788 KB631815 AEE61750.1 ERL86438.1 LJIJ01006756 ODM86922.1 GAMC01000092 JAC06464.1 CH916368 EDW03482.1 CH964182 EDW80268.1 AJWK01010790 AJWK01010791 AJWK01010792 DS232359 EDS40602.1 JTDY01006277 KOB66148.1 CP012523 ALC40109.1 GEFM01000699 JAP75097.1 GFWV01018976 MAA43704.1 GANP01000875 JAB83593.1 GADI01005966 JAA67842.1 CH933807 EDW13031.1 GFDF01001647 JAV12437.1 GETE01000079 JAT79266.1 ABJB010110070 ABJB010413306 ABJB010704850 DS748754 EEC07870.1 GEFH01000055 JAP68526.1 GFTR01002439 JAW13987.1 GFPF01010121 MAA21267.1 GBGD01003379 JAC85510.1 JO843996 AEO35613.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
+ More
UP000075884 UP000027135 UP000069940 UP000249989 UP000075885 UP000075886 UP000076407 UP000007062 UP000075840 UP000235965 UP000008820 UP000000673 UP000095301 UP000075920 UP000075880 UP000075883 UP000030765 UP000092445 UP000092444 UP000076408 UP000075901 UP000069272 UP000192221 UP000037069 UP000008711 UP000000803 UP000075881 UP000078200 UP000198287 UP000002282 UP000092460 UP000092443 UP000001292 UP000094527 UP000007801 UP000091820 UP000075900 UP000095300 UP000268350 UP000183832 UP000008744 UP000001819 UP000030742 UP000001070 UP000007798 UP000192223 UP000092461 UP000002320 UP000079169 UP000037510 UP000092553 UP000009192 UP000001555
UP000075884 UP000027135 UP000069940 UP000249989 UP000075885 UP000075886 UP000076407 UP000007062 UP000075840 UP000235965 UP000008820 UP000000673 UP000095301 UP000075920 UP000075880 UP000075883 UP000030765 UP000092445 UP000092444 UP000076408 UP000075901 UP000069272 UP000192221 UP000037069 UP000008711 UP000000803 UP000075881 UP000078200 UP000198287 UP000002282 UP000092460 UP000092443 UP000001292 UP000094527 UP000007801 UP000091820 UP000075900 UP000095300 UP000268350 UP000183832 UP000008744 UP000001819 UP000030742 UP000001070 UP000007798 UP000192223 UP000092461 UP000002320 UP000079169 UP000037510 UP000092553 UP000009192 UP000001555
SUPFAM
SSF109925
SSF109925
ProteinModelPortal
H9J226
A0A1E1WKG1
A0A1E1WGE6
A0A2W1BPS5
A0A2A4K456
A0A212FEZ7
+ More
A0A2H1V1S6 A0A0N1IB01 A0A194PNQ7 V5G9Y9 D6WAK3 A0A182MYC3 A0A067R041 A0A182GYX2 A0A023EFC6 A0A0K8TQA0 A0A182PTJ9 A0A182QM87 A0A1S4F7K0 A0A182WUS0 A0NDZ8 A0A182HFT0 A0A2J7PC16 Q17CW4 W5JI25 T1PI10 A0A182VTA5 A0A182JFP3 A0A182MC23 A0A084VKV8 A0A1A9ZDD4 A0A1B0F9P0 A0A2M3ZJM3 A0A0P6J945 A0A2M4C2W0 A0A2M4C2F7 A0A182YA21 A0A182SAY7 A0A182FUJ0 A0A1W4UW53 A0A0L0C5X9 A0A0J9R6D0 B3NKT6 M9NEW0 A0A2M4AWA9 A0A182JXL6 A0A1A9V9S9 A0A226F1Y1 A0A0P5R385 B4P6Y7 A0A1Q3FLC0 A0A1B0B7P6 A0A1A9XL35 B4IFL6 A0A0N8DNU6 A0A1D2MPN6 A0A0P9A3Q8 A0A1Y1N7Z6 A0A1Q3FLI5 A0A0P5ZP51 A0A1A9WKT7 A0A182RLQ9 A0A034WHS5 A0A0C9RLB6 A0A1I8QBF6 A0A2R5LHK4 A0A3B0KEX3 A0A1L8EBA6 A0A1L8EC29 A0A1J1IVS6 A0A0K8U1R0 B4GJJ4 B5DHS6 J3JU75 A0A1D2M1V2 W8C876 B4JDW2 B4N7C7 A0A1W4X6C5 A0A1B0CG69 B0X4Y7 A0A1S3D3I4 A0A0L7KSC4 A0A0M3QU68 A0A131Y8F5 A0A293MS95 V5I4X2 A0A0K8R9J7 B4KF48 A0A1L8E161 A0A1D2AJ99 B7PMP8 A0A131XS76 A0A224XNC1 A0A224YUI2 A0A069DNV6 G3MQ45
A0A2H1V1S6 A0A0N1IB01 A0A194PNQ7 V5G9Y9 D6WAK3 A0A182MYC3 A0A067R041 A0A182GYX2 A0A023EFC6 A0A0K8TQA0 A0A182PTJ9 A0A182QM87 A0A1S4F7K0 A0A182WUS0 A0NDZ8 A0A182HFT0 A0A2J7PC16 Q17CW4 W5JI25 T1PI10 A0A182VTA5 A0A182JFP3 A0A182MC23 A0A084VKV8 A0A1A9ZDD4 A0A1B0F9P0 A0A2M3ZJM3 A0A0P6J945 A0A2M4C2W0 A0A2M4C2F7 A0A182YA21 A0A182SAY7 A0A182FUJ0 A0A1W4UW53 A0A0L0C5X9 A0A0J9R6D0 B3NKT6 M9NEW0 A0A2M4AWA9 A0A182JXL6 A0A1A9V9S9 A0A226F1Y1 A0A0P5R385 B4P6Y7 A0A1Q3FLC0 A0A1B0B7P6 A0A1A9XL35 B4IFL6 A0A0N8DNU6 A0A1D2MPN6 A0A0P9A3Q8 A0A1Y1N7Z6 A0A1Q3FLI5 A0A0P5ZP51 A0A1A9WKT7 A0A182RLQ9 A0A034WHS5 A0A0C9RLB6 A0A1I8QBF6 A0A2R5LHK4 A0A3B0KEX3 A0A1L8EBA6 A0A1L8EC29 A0A1J1IVS6 A0A0K8U1R0 B4GJJ4 B5DHS6 J3JU75 A0A1D2M1V2 W8C876 B4JDW2 B4N7C7 A0A1W4X6C5 A0A1B0CG69 B0X4Y7 A0A1S3D3I4 A0A0L7KSC4 A0A0M3QU68 A0A131Y8F5 A0A293MS95 V5I4X2 A0A0K8R9J7 B4KF48 A0A1L8E161 A0A1D2AJ99 B7PMP8 A0A131XS76 A0A224XNC1 A0A224YUI2 A0A069DNV6 G3MQ45
Ontologies
GO
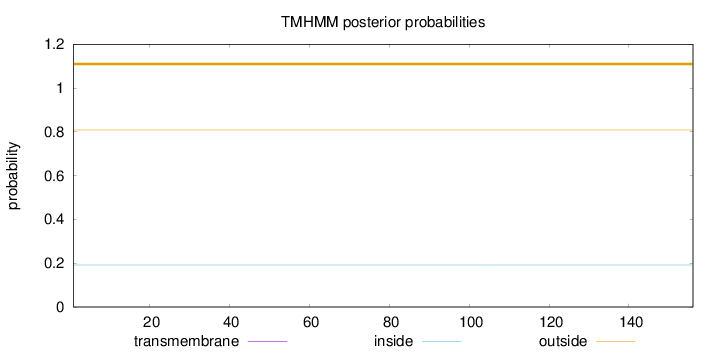
Topology
Length:
156
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9e-05
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.19215
outside
1 - 156
Population Genetic Test Statistics
Pi
158.69454
Theta
151.069755
Tajima's D
-0.861468
CLR
207.605376
CSRT
0.159542022898855
Interpretation
Uncertain
