Pre Gene Modal
BGIBMGA003632
Annotation
PREDICTED:_uncharacterized_protein_C45G9.7_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 2.496
Sequence
CDS
ATGGCGAAGATGGCATTTCAACACCAGGCTGGAACAGCTATGGAGTGTCTAAGTATACCGATAACTTTACAAAAAGAGGCTGGTGTTGACCCAGATGGAAGAGAAGTAATGAAATGTGGTTTTAAAATAGGAGGAGGCATTGATCAAGACTTTCGTAAGAGCCCCCAGGGATATACAGATAACGGTATTTATGTAACTGAAGTTCATGAAGGAAGTCCAGCAGCAAAATCAGGTTTAAGAATGCATGATAAAATACTACAGTGCAATGGTTATGACTTTACAATGGTAACTCACAAAAAAGCAGTTAGTTATATTAAGAAACACCCTATTCTTAACTTGCTTGTGGCCAGAAAAGGAGTTACATCTACATAA
Protein
MAKMAFQHQAGTAMECLSIPITLQKEAGVDPDGREVMKCGFKIGGGIDQDFRKSPQGYTDNGIYVTEVHEGSPAAKSGLRMHDKILQCNGYDFTMVTHKKAVSYIKKHPILNLLVARKGVTST
Summary
Uniprot
H9J295
A0A2W1BPM8
A0A2H1V0G1
A0A1E1WBQ6
A0A2A4K4L3
A0A194PVA7
+ More
S4NZC0 A0A212FF26 A0A1B6FN55 A0A1B6H6T0 A0A1B6DVK8 A0A1B6KNH2 E0VQ23 A0A224Y0B7 A0A0P4VLJ0 A0A0V0GDN9 A0A023FAN8 A0A069DVT6 A0A2P8XK57 A0A0A9XU66 R4WJF4 A0A0L7R8B7 T1HCA3 A0A182J1Y6 A0A1Q3FBH0 B0WLS2 Q0IFI2 A0A182GZG9 A0A2J7R2Q6 A0A2A3EIB9 A0A0M8ZPU3 A0A088APZ0 A0A182Q7W4 A0A1L8E3J6 A0A182Y181 A0A1B0DG37 A0A182MNQ9 A0A182PHK2 A0A182UWL8 A0A182NGU1 A0A182WB28 A0A182U0H0 A0A084W9Z8 A0A182LJ74 Q7Q6X0 A0A182RRV1 A0A182HWU7 A0A1B0EWU7 A0A182XMZ1 A0A3L8DC54 A0A158NJ40 E2BK51 A0A2M3ZDV4 A0A2M4AX90 W5JFJ9 E1ZVL3 A0A182FDM7 A0A2M4C3L2 A0A0K8TMY7 A0A232FJY4 A0A1I8MWU5 A0A0L0CLU5 F4W795 A0A2H8TW41 J9JQJ3 A0A2S2P5E9 A0A1W4VRI2 B4LGL0 A0A0J9RMQ4 B4L0A4 A0A3B0KLP4 Q29DV8 B4PCL4 B3M509 B4MLB4 B4HVH9 B4J2S8 B3NER8 Q9W0P4 A0A1S3DKI2 A0A2S2Q8H0 A0A1A9WS90 A0A1I8Q380 A0A336LGY3 A0A1B0FEQ6 A0A1A9YH00 A0A1A9UG32 A0A1B0BC65 A0A1A9ZKF7 T1EA85 K7IVB3 A0A0C9PVZ9 A0A0A1X0T1 A0A0K8UQK5 A0A0N1IHB6 A0A1Y1L5R6 A0A0T6AUL3 D6W7Z1 V5H9M5 B7PJR1
S4NZC0 A0A212FF26 A0A1B6FN55 A0A1B6H6T0 A0A1B6DVK8 A0A1B6KNH2 E0VQ23 A0A224Y0B7 A0A0P4VLJ0 A0A0V0GDN9 A0A023FAN8 A0A069DVT6 A0A2P8XK57 A0A0A9XU66 R4WJF4 A0A0L7R8B7 T1HCA3 A0A182J1Y6 A0A1Q3FBH0 B0WLS2 Q0IFI2 A0A182GZG9 A0A2J7R2Q6 A0A2A3EIB9 A0A0M8ZPU3 A0A088APZ0 A0A182Q7W4 A0A1L8E3J6 A0A182Y181 A0A1B0DG37 A0A182MNQ9 A0A182PHK2 A0A182UWL8 A0A182NGU1 A0A182WB28 A0A182U0H0 A0A084W9Z8 A0A182LJ74 Q7Q6X0 A0A182RRV1 A0A182HWU7 A0A1B0EWU7 A0A182XMZ1 A0A3L8DC54 A0A158NJ40 E2BK51 A0A2M3ZDV4 A0A2M4AX90 W5JFJ9 E1ZVL3 A0A182FDM7 A0A2M4C3L2 A0A0K8TMY7 A0A232FJY4 A0A1I8MWU5 A0A0L0CLU5 F4W795 A0A2H8TW41 J9JQJ3 A0A2S2P5E9 A0A1W4VRI2 B4LGL0 A0A0J9RMQ4 B4L0A4 A0A3B0KLP4 Q29DV8 B4PCL4 B3M509 B4MLB4 B4HVH9 B4J2S8 B3NER8 Q9W0P4 A0A1S3DKI2 A0A2S2Q8H0 A0A1A9WS90 A0A1I8Q380 A0A336LGY3 A0A1B0FEQ6 A0A1A9YH00 A0A1A9UG32 A0A1B0BC65 A0A1A9ZKF7 T1EA85 K7IVB3 A0A0C9PVZ9 A0A0A1X0T1 A0A0K8UQK5 A0A0N1IHB6 A0A1Y1L5R6 A0A0T6AUL3 D6W7Z1 V5H9M5 B7PJR1
Pubmed
19121390
28756777
26354079
23622113
22118469
20566863
+ More
27129103 25474469 26334808 29403074 25401762 26823975 23691247 17510324 26483478 25244985 24438588 20966253 12364791 14747013 17210077 30249741 21347285 20798317 20920257 23761445 26369729 28648823 25315136 26108605 21719571 17994087 22936249 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 25830018 28004739 18362917 19820115 25765539
27129103 25474469 26334808 29403074 25401762 26823975 23691247 17510324 26483478 25244985 24438588 20966253 12364791 14747013 17210077 30249741 21347285 20798317 20920257 23761445 26369729 28648823 25315136 26108605 21719571 17994087 22936249 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 25830018 28004739 18362917 19820115 25765539
EMBL
BABH01007533
BABH01007534
KZ149992
PZC75564.1
ODYU01000103
SOQ34301.1
+ More
GDQN01006687 JAT84367.1 NWSH01000167 PCG78848.1 KQ459597 KPI95060.1 GAIX01013455 JAA79105.1 AGBW02008872 OWR52320.1 GECZ01018178 JAS51591.1 GECU01037328 JAS70378.1 GEDC01007615 JAS29683.1 GEBQ01027000 JAT12977.1 DS235389 EEB15479.1 GFTR01002084 JAW14342.1 GDKW01000602 JAI55993.1 GECL01000069 JAP06055.1 GBBI01000175 JAC18537.1 GBGD01003485 JAC85404.1 PYGN01001874 PSN32375.1 GBHO01022929 GBRD01014379 GDHC01017203 GDHC01008714 JAG20675.1 JAG51447.1 JAQ01426.1 JAQ09915.1 AK417660 BAN20875.1 KQ414632 KOC67127.1 ACPB03007695 GFDL01010202 JAV24843.1 DS231990 EDS30650.1 CH477317 EAT43730.1 EAT43732.1 JXUM01099623 KQ564506 KXJ72169.1 NEVH01007828 PNF35117.1 KZ288232 PBC31535.1 KQ435922 KOX68635.1 AXCN02000366 GFDF01000783 JAV13301.1 AJVK01059230 AXCM01007282 ATLV01021973 KE525326 KFB47042.1 AAAB01008960 EAA11322.2 APCN01001540 AJWK01010315 QOIP01000010 RLU18100.1 ADTU01017434 GL448740 EFN83959.1 GGFM01005869 MBW26620.1 GGFK01012095 MBW45416.1 ADMH02001639 ETN61655.1 GL434548 EFN74782.1 GGFJ01010751 MBW59892.1 GDAI01001876 JAI15727.1 NNAY01000100 OXU30985.1 JRES01000215 KNC33236.1 GL887813 EGI69979.1 GFXV01006324 MBW18129.1 ABLF02037161 GGMR01011507 MBY24126.1 CH940647 EDW69448.1 CM002912 KMY96699.1 CH933809 EDW18050.1 OUUW01000012 SPP87499.1 CH379070 EAL30305.1 CM000159 EDW92737.1 CH902618 EDV39488.1 CH963847 EDW73372.1 CH480817 EDW49944.1 CH916366 EDV96069.1 CH954178 EDV50063.1 AE014296 AY069359 KX531760 AAF47399.1 AAL39504.1 ANY27570.1 GGMS01004834 MBY74037.1 UFQT01000004 SSX17252.1 CCAG010020479 JXJN01011858 GAMD01000657 JAB00934.1 AAZX01016693 AAZX01019382 GBYB01005658 JAG75425.1 GBXI01009353 GBXI01001180 JAD04939.1 JAD13112.1 GDHF01027521 GDHF01023541 JAI24793.1 JAI28773.1 KQ460226 KPJ16466.1 GEZM01070134 JAV66437.1 LJIG01022763 KRT78847.1 KQ971307 EFA11159.2 GANP01004493 JAB79975.1 ABJB010002302 ABJB011039511 DS727794 EEC06833.1
GDQN01006687 JAT84367.1 NWSH01000167 PCG78848.1 KQ459597 KPI95060.1 GAIX01013455 JAA79105.1 AGBW02008872 OWR52320.1 GECZ01018178 JAS51591.1 GECU01037328 JAS70378.1 GEDC01007615 JAS29683.1 GEBQ01027000 JAT12977.1 DS235389 EEB15479.1 GFTR01002084 JAW14342.1 GDKW01000602 JAI55993.1 GECL01000069 JAP06055.1 GBBI01000175 JAC18537.1 GBGD01003485 JAC85404.1 PYGN01001874 PSN32375.1 GBHO01022929 GBRD01014379 GDHC01017203 GDHC01008714 JAG20675.1 JAG51447.1 JAQ01426.1 JAQ09915.1 AK417660 BAN20875.1 KQ414632 KOC67127.1 ACPB03007695 GFDL01010202 JAV24843.1 DS231990 EDS30650.1 CH477317 EAT43730.1 EAT43732.1 JXUM01099623 KQ564506 KXJ72169.1 NEVH01007828 PNF35117.1 KZ288232 PBC31535.1 KQ435922 KOX68635.1 AXCN02000366 GFDF01000783 JAV13301.1 AJVK01059230 AXCM01007282 ATLV01021973 KE525326 KFB47042.1 AAAB01008960 EAA11322.2 APCN01001540 AJWK01010315 QOIP01000010 RLU18100.1 ADTU01017434 GL448740 EFN83959.1 GGFM01005869 MBW26620.1 GGFK01012095 MBW45416.1 ADMH02001639 ETN61655.1 GL434548 EFN74782.1 GGFJ01010751 MBW59892.1 GDAI01001876 JAI15727.1 NNAY01000100 OXU30985.1 JRES01000215 KNC33236.1 GL887813 EGI69979.1 GFXV01006324 MBW18129.1 ABLF02037161 GGMR01011507 MBY24126.1 CH940647 EDW69448.1 CM002912 KMY96699.1 CH933809 EDW18050.1 OUUW01000012 SPP87499.1 CH379070 EAL30305.1 CM000159 EDW92737.1 CH902618 EDV39488.1 CH963847 EDW73372.1 CH480817 EDW49944.1 CH916366 EDV96069.1 CH954178 EDV50063.1 AE014296 AY069359 KX531760 AAF47399.1 AAL39504.1 ANY27570.1 GGMS01004834 MBY74037.1 UFQT01000004 SSX17252.1 CCAG010020479 JXJN01011858 GAMD01000657 JAB00934.1 AAZX01016693 AAZX01019382 GBYB01005658 JAG75425.1 GBXI01009353 GBXI01001180 JAD04939.1 JAD13112.1 GDHF01027521 GDHF01023541 JAI24793.1 JAI28773.1 KQ460226 KPJ16466.1 GEZM01070134 JAV66437.1 LJIG01022763 KRT78847.1 KQ971307 EFA11159.2 GANP01004493 JAB79975.1 ABJB010002302 ABJB011039511 DS727794 EEC06833.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000009046
UP000245037
+ More
UP000053825 UP000015103 UP000075880 UP000002320 UP000008820 UP000069940 UP000249989 UP000235965 UP000242457 UP000053105 UP000005203 UP000075886 UP000076408 UP000092462 UP000075883 UP000075885 UP000075903 UP000075884 UP000075920 UP000075902 UP000030765 UP000075882 UP000007062 UP000075900 UP000075840 UP000092461 UP000076407 UP000279307 UP000005205 UP000008237 UP000000673 UP000000311 UP000069272 UP000215335 UP000095301 UP000037069 UP000007755 UP000007819 UP000192221 UP000008792 UP000009192 UP000268350 UP000001819 UP000002282 UP000007801 UP000007798 UP000001292 UP000001070 UP000008711 UP000000803 UP000079169 UP000091820 UP000095300 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000002358 UP000053240 UP000007266 UP000001555
UP000053825 UP000015103 UP000075880 UP000002320 UP000008820 UP000069940 UP000249989 UP000235965 UP000242457 UP000053105 UP000005203 UP000075886 UP000076408 UP000092462 UP000075883 UP000075885 UP000075903 UP000075884 UP000075920 UP000075902 UP000030765 UP000075882 UP000007062 UP000075900 UP000075840 UP000092461 UP000076407 UP000279307 UP000005205 UP000008237 UP000000673 UP000000311 UP000069272 UP000215335 UP000095301 UP000037069 UP000007755 UP000007819 UP000192221 UP000008792 UP000009192 UP000268350 UP000001819 UP000002282 UP000007801 UP000007798 UP000001292 UP000001070 UP000008711 UP000000803 UP000079169 UP000091820 UP000095300 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000002358 UP000053240 UP000007266 UP000001555
Pfam
PF00595 PDZ
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
H9J295
A0A2W1BPM8
A0A2H1V0G1
A0A1E1WBQ6
A0A2A4K4L3
A0A194PVA7
+ More
S4NZC0 A0A212FF26 A0A1B6FN55 A0A1B6H6T0 A0A1B6DVK8 A0A1B6KNH2 E0VQ23 A0A224Y0B7 A0A0P4VLJ0 A0A0V0GDN9 A0A023FAN8 A0A069DVT6 A0A2P8XK57 A0A0A9XU66 R4WJF4 A0A0L7R8B7 T1HCA3 A0A182J1Y6 A0A1Q3FBH0 B0WLS2 Q0IFI2 A0A182GZG9 A0A2J7R2Q6 A0A2A3EIB9 A0A0M8ZPU3 A0A088APZ0 A0A182Q7W4 A0A1L8E3J6 A0A182Y181 A0A1B0DG37 A0A182MNQ9 A0A182PHK2 A0A182UWL8 A0A182NGU1 A0A182WB28 A0A182U0H0 A0A084W9Z8 A0A182LJ74 Q7Q6X0 A0A182RRV1 A0A182HWU7 A0A1B0EWU7 A0A182XMZ1 A0A3L8DC54 A0A158NJ40 E2BK51 A0A2M3ZDV4 A0A2M4AX90 W5JFJ9 E1ZVL3 A0A182FDM7 A0A2M4C3L2 A0A0K8TMY7 A0A232FJY4 A0A1I8MWU5 A0A0L0CLU5 F4W795 A0A2H8TW41 J9JQJ3 A0A2S2P5E9 A0A1W4VRI2 B4LGL0 A0A0J9RMQ4 B4L0A4 A0A3B0KLP4 Q29DV8 B4PCL4 B3M509 B4MLB4 B4HVH9 B4J2S8 B3NER8 Q9W0P4 A0A1S3DKI2 A0A2S2Q8H0 A0A1A9WS90 A0A1I8Q380 A0A336LGY3 A0A1B0FEQ6 A0A1A9YH00 A0A1A9UG32 A0A1B0BC65 A0A1A9ZKF7 T1EA85 K7IVB3 A0A0C9PVZ9 A0A0A1X0T1 A0A0K8UQK5 A0A0N1IHB6 A0A1Y1L5R6 A0A0T6AUL3 D6W7Z1 V5H9M5 B7PJR1
S4NZC0 A0A212FF26 A0A1B6FN55 A0A1B6H6T0 A0A1B6DVK8 A0A1B6KNH2 E0VQ23 A0A224Y0B7 A0A0P4VLJ0 A0A0V0GDN9 A0A023FAN8 A0A069DVT6 A0A2P8XK57 A0A0A9XU66 R4WJF4 A0A0L7R8B7 T1HCA3 A0A182J1Y6 A0A1Q3FBH0 B0WLS2 Q0IFI2 A0A182GZG9 A0A2J7R2Q6 A0A2A3EIB9 A0A0M8ZPU3 A0A088APZ0 A0A182Q7W4 A0A1L8E3J6 A0A182Y181 A0A1B0DG37 A0A182MNQ9 A0A182PHK2 A0A182UWL8 A0A182NGU1 A0A182WB28 A0A182U0H0 A0A084W9Z8 A0A182LJ74 Q7Q6X0 A0A182RRV1 A0A182HWU7 A0A1B0EWU7 A0A182XMZ1 A0A3L8DC54 A0A158NJ40 E2BK51 A0A2M3ZDV4 A0A2M4AX90 W5JFJ9 E1ZVL3 A0A182FDM7 A0A2M4C3L2 A0A0K8TMY7 A0A232FJY4 A0A1I8MWU5 A0A0L0CLU5 F4W795 A0A2H8TW41 J9JQJ3 A0A2S2P5E9 A0A1W4VRI2 B4LGL0 A0A0J9RMQ4 B4L0A4 A0A3B0KLP4 Q29DV8 B4PCL4 B3M509 B4MLB4 B4HVH9 B4J2S8 B3NER8 Q9W0P4 A0A1S3DKI2 A0A2S2Q8H0 A0A1A9WS90 A0A1I8Q380 A0A336LGY3 A0A1B0FEQ6 A0A1A9YH00 A0A1A9UG32 A0A1B0BC65 A0A1A9ZKF7 T1EA85 K7IVB3 A0A0C9PVZ9 A0A0A1X0T1 A0A0K8UQK5 A0A0N1IHB6 A0A1Y1L5R6 A0A0T6AUL3 D6W7Z1 V5H9M5 B7PJR1
PDB
2VZ5
E-value=7.02868e-16,
Score=198
Ontologies
GO
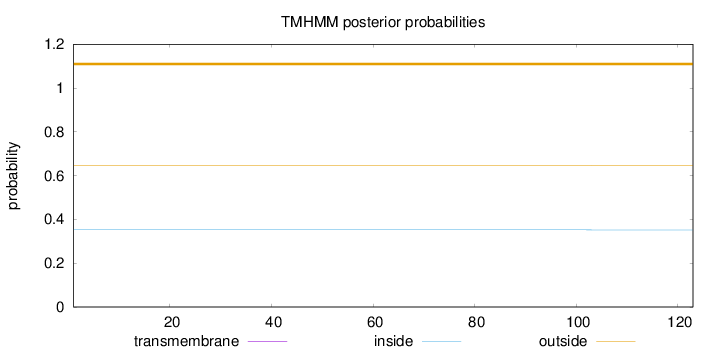
Topology
Length:
123
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00116
Exp number, first 60 AAs:
0.00052
Total prob of N-in:
0.35225
outside
1 - 123
Population Genetic Test Statistics
Pi
149.317133
Theta
183.026916
Tajima's D
-0.579523
CLR
14.495959
CSRT
0.229188540572971
Interpretation
Uncertain
