Gene
KWMTBOMO02941 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003562
Annotation
NADH_dehydrogenase_1_alpha_subcomplex_subunit_5_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.114
Sequence
CDS
ATGGGTTTATTAAAAAAGACAACAGGTCTAACAGGATTGGCAGTCGCCGCAAATCCGCATCACACTTTGGGCGCCCTATATGGTAAAATCTTACGTACTCTCCAAAAAATGCCAGAAACATCAGTTTATCGTAAATATACGGAACAAATAGTACGTGAAAGAGCAGCGGTTTTAACTCAGACTAAAGACAGCTTTGAAATCGAAAAGAAAATCAACTGCGGTCAAGCTGAGGAATTGATAATTCAAGCTGAAAATGAATTAAACCTAGCTAGGAAGATGTTAAATTGGAAGCCGTGGGAACCACTAATGGCTAAGCCTCCTAAAGGTCAATGGGAATGGCCTCCATCTAAAGCTTAA
Protein
MGLLKKTTGLTGLAVAANPHHTLGALYGKILRTLQKMPETSVYRKYTEQIVRERAAVLTQTKDSFEIEKKINCGQAEELIIQAENELNLARKMLNWKPWEPLMAKPPKGQWEWPPSKA
Summary
Uniprot
H9J225
Q1G0N4
A0A0L7KSC1
A0A212FEZ8
A0A0N1IIK2
S4NXE9
+ More
I4DK45 A0A2W1BUY1 A0A2A4K3L6 A0A1E1WDB2 A0A2H1V0C5 A0A1B0CRS4 A0A1B0CJQ6 A0A2P8XLU3 A0A2J7R2Q5 A0A1L8E0G1 A0A1Y1MUQ6 D6X292 A0A182Y179 A0A182LJ73 A0A182QLJ1 V5GFE4 A0A182SZT9 A0A0K8TSU9 Q1HQT0 A0A2M4CNF4 A0A1Q3FKX8 A0A182FDM6 B0WLS3 A0A023EFW7 A0A2H1WMG4 A0A2M3Z686 A0A182RRV3 A0A182GZH1 A0A084VCA6 B4LB56 A0A182VB56 A0A182UDI7 A0A182XMZ3 Q7PNP3 A0A182HWU9 A0A2M4C509 A0A2M4C4M6 T1DJH3 A0A182WB26 A0A1S4F8R1 A0A2M4C3B0 Q0IFI1 C4WUD5 W5JFR1 A0A1A9WTA2 A0A2S2P7F5 J9K5T5 A0A2M4AFK0 A0A067RC26 A0A2M4AFZ0 A0A1I8QB73 A0A182PHK0 A0A1A9Y053 B4L9G6 A0A1A9UG23 A0A182JBP1 A0A1I8M7P5 A0A2H8TJC9 B4PEG9 Q9VTB4 B4HDR3 B4QP91 B3NCN0 A0A0A1WEU3 A0A1W4UV17 A0A1L8EEP4 A0A1D2MW47 A0A0P5F2H7 A0A0P4XNG0 A0A0P5AFP3 Q29E06 B4H439 A0A0P5HUK1 A0A0P5YA62 A0A0N8D3P7 A0A0P5HQ27 A0A0N8DB15 A0A0P6D5H1 A0A0P6D7H0 A0A0P6H4W8 A0A0P5D8A8 A0A293M0Q4 A0A1L8EF84 A0A182NGT9 A0A1E1X0T2 W8C0Z7 A0A0M3QW54 A0A0P5TXP9 A0A034WVB1 B4MLV5 A0A0N8DM33 A0A1B6KJS4 A0A224YWE1 A0A224Z4Q8
I4DK45 A0A2W1BUY1 A0A2A4K3L6 A0A1E1WDB2 A0A2H1V0C5 A0A1B0CRS4 A0A1B0CJQ6 A0A2P8XLU3 A0A2J7R2Q5 A0A1L8E0G1 A0A1Y1MUQ6 D6X292 A0A182Y179 A0A182LJ73 A0A182QLJ1 V5GFE4 A0A182SZT9 A0A0K8TSU9 Q1HQT0 A0A2M4CNF4 A0A1Q3FKX8 A0A182FDM6 B0WLS3 A0A023EFW7 A0A2H1WMG4 A0A2M3Z686 A0A182RRV3 A0A182GZH1 A0A084VCA6 B4LB56 A0A182VB56 A0A182UDI7 A0A182XMZ3 Q7PNP3 A0A182HWU9 A0A2M4C509 A0A2M4C4M6 T1DJH3 A0A182WB26 A0A1S4F8R1 A0A2M4C3B0 Q0IFI1 C4WUD5 W5JFR1 A0A1A9WTA2 A0A2S2P7F5 J9K5T5 A0A2M4AFK0 A0A067RC26 A0A2M4AFZ0 A0A1I8QB73 A0A182PHK0 A0A1A9Y053 B4L9G6 A0A1A9UG23 A0A182JBP1 A0A1I8M7P5 A0A2H8TJC9 B4PEG9 Q9VTB4 B4HDR3 B4QP91 B3NCN0 A0A0A1WEU3 A0A1W4UV17 A0A1L8EEP4 A0A1D2MW47 A0A0P5F2H7 A0A0P4XNG0 A0A0P5AFP3 Q29E06 B4H439 A0A0P5HUK1 A0A0P5YA62 A0A0N8D3P7 A0A0P5HQ27 A0A0N8DB15 A0A0P6D5H1 A0A0P6D7H0 A0A0P6H4W8 A0A0P5D8A8 A0A293M0Q4 A0A1L8EF84 A0A182NGT9 A0A1E1X0T2 W8C0Z7 A0A0M3QW54 A0A0P5TXP9 A0A034WVB1 B4MLV5 A0A0N8DM33 A0A1B6KJS4 A0A224YWE1 A0A224Z4Q8
Pubmed
19121390
26227816
22118469
26354079
23622113
22651552
+ More
28756777 29403074 28004739 18362917 19820115 25244985 20966253 26369729 17204158 24945155 26483478 24438588 17994087 12364791 14747013 17210077 17510324 20920257 23761445 24845553 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 27289101 15632085 28503490 24495485 25348373 28797301
28756777 29403074 28004739 18362917 19820115 25244985 20966253 26369729 17204158 24945155 26483478 24438588 17994087 12364791 14747013 17210077 17510324 20920257 23761445 24845553 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 27289101 15632085 28503490 24495485 25348373 28797301
EMBL
BABH01007534
DQ534194
ABF60226.1
JTDY01006277
KOB66147.1
AGBW02008872
+ More
OWR52321.1 KQ460226 KPJ16465.1 GAIX01009059 JAA83501.1 AK401663 KQ459597 BAM18285.1 KPI95059.1 KZ149992 PZC75563.1 NWSH01000167 PCG78847.1 GDQN01006098 JAT84956.1 ODYU01000103 SOQ34300.1 AJWK01025249 AJWK01014849 PYGN01001758 PSN32971.1 NEVH01007828 PNF35119.1 GFDF01001864 JAV12220.1 GEZM01020392 JAV89279.1 KQ971371 EFA10821.1 AXCN02000366 GALX01005772 JAB62694.1 GDAI01000166 JAI17437.1 DQ440364 ABF18397.1 GGFL01002684 MBW66862.1 GFDL01006891 JAV28154.1 DS231990 EDS30651.1 GAPW01005847 JAC07751.1 ODYU01009642 SOQ54218.1 GGFM01003282 MBW24033.1 JXUM01099624 KQ564506 KXJ72171.1 ATLV01010628 KE524589 KFB35600.1 CH940647 EDW68620.1 AAAB01008960 EAA11832.4 APCN01001541 GGFJ01010927 MBW60068.1 GGFJ01010950 MBW60091.1 GAMD01001446 JAB00145.1 GGFJ01010669 MBW59810.1 CH477317 EAT43733.1 AK341020 BAH71505.1 ADMH02001639 ETN61654.1 GGMR01012754 MBY25373.1 ABLF02021646 GGFK01006248 MBW39569.1 KK852744 KDR17347.1 GGFK01006227 MBW39548.1 CH933816 EDW17341.1 GFXV01001593 MBW13398.1 CM000159 EDW94035.1 AE014296 AY070675 AAF50138.2 AAL48146.1 CH480815 EDW41003.1 CM000363 CM002912 EDX09989.1 KMY98872.1 CH954178 EDV51327.1 GBXI01017302 JAC96989.1 GFDG01001607 JAV17192.1 LJIJ01000461 ODM97226.1 GDIQ01263962 JAJ87762.1 GDIP01248947 LRGB01003123 JAI74454.1 KZS04504.1 GDIP01212906 JAJ10496.1 CH379070 EAL30257.2 CH479208 EDW31154.1 GDIQ01222071 JAK29654.1 GDIP01062172 JAM41543.1 GDIP01071871 JAM31844.1 GDIQ01224671 JAK27054.1 GDIP01051327 JAM52388.1 GDIQ01084671 JAN10066.1 GDIQ01081671 JAN13066.1 GDIQ01024431 JAN70306.1 GDIP01161478 JAJ61924.1 GFWV01008932 MAA33661.1 GFDG01001606 JAV17193.1 GFAC01006309 JAT92879.1 GAMC01001203 JAC05353.1 CP012525 ALC43553.1 GDIP01120563 JAL83151.1 GAKP01000373 JAC58579.1 CH963847 EDW73166.1 GDIP01020383 JAM83332.1 GEBQ01028274 JAT11703.1 GFPF01010099 MAA21245.1 GFPF01010098 MAA21244.1
OWR52321.1 KQ460226 KPJ16465.1 GAIX01009059 JAA83501.1 AK401663 KQ459597 BAM18285.1 KPI95059.1 KZ149992 PZC75563.1 NWSH01000167 PCG78847.1 GDQN01006098 JAT84956.1 ODYU01000103 SOQ34300.1 AJWK01025249 AJWK01014849 PYGN01001758 PSN32971.1 NEVH01007828 PNF35119.1 GFDF01001864 JAV12220.1 GEZM01020392 JAV89279.1 KQ971371 EFA10821.1 AXCN02000366 GALX01005772 JAB62694.1 GDAI01000166 JAI17437.1 DQ440364 ABF18397.1 GGFL01002684 MBW66862.1 GFDL01006891 JAV28154.1 DS231990 EDS30651.1 GAPW01005847 JAC07751.1 ODYU01009642 SOQ54218.1 GGFM01003282 MBW24033.1 JXUM01099624 KQ564506 KXJ72171.1 ATLV01010628 KE524589 KFB35600.1 CH940647 EDW68620.1 AAAB01008960 EAA11832.4 APCN01001541 GGFJ01010927 MBW60068.1 GGFJ01010950 MBW60091.1 GAMD01001446 JAB00145.1 GGFJ01010669 MBW59810.1 CH477317 EAT43733.1 AK341020 BAH71505.1 ADMH02001639 ETN61654.1 GGMR01012754 MBY25373.1 ABLF02021646 GGFK01006248 MBW39569.1 KK852744 KDR17347.1 GGFK01006227 MBW39548.1 CH933816 EDW17341.1 GFXV01001593 MBW13398.1 CM000159 EDW94035.1 AE014296 AY070675 AAF50138.2 AAL48146.1 CH480815 EDW41003.1 CM000363 CM002912 EDX09989.1 KMY98872.1 CH954178 EDV51327.1 GBXI01017302 JAC96989.1 GFDG01001607 JAV17192.1 LJIJ01000461 ODM97226.1 GDIQ01263962 JAJ87762.1 GDIP01248947 LRGB01003123 JAI74454.1 KZS04504.1 GDIP01212906 JAJ10496.1 CH379070 EAL30257.2 CH479208 EDW31154.1 GDIQ01222071 JAK29654.1 GDIP01062172 JAM41543.1 GDIP01071871 JAM31844.1 GDIQ01224671 JAK27054.1 GDIP01051327 JAM52388.1 GDIQ01084671 JAN10066.1 GDIQ01081671 JAN13066.1 GDIQ01024431 JAN70306.1 GDIP01161478 JAJ61924.1 GFWV01008932 MAA33661.1 GFDG01001606 JAV17193.1 GFAC01006309 JAT92879.1 GAMC01001203 JAC05353.1 CP012525 ALC43553.1 GDIP01120563 JAL83151.1 GAKP01000373 JAC58579.1 CH963847 EDW73166.1 GDIP01020383 JAM83332.1 GEBQ01028274 JAT11703.1 GFPF01010099 MAA21245.1 GFPF01010098 MAA21244.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000092461 UP000245037 UP000235965 UP000007266 UP000076408 UP000075882 UP000075886 UP000075901 UP000069272 UP000002320 UP000075900 UP000069940 UP000249989 UP000030765 UP000008792 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000075920 UP000008820 UP000000673 UP000091820 UP000007819 UP000027135 UP000095300 UP000075885 UP000092443 UP000009192 UP000078200 UP000075880 UP000095301 UP000002282 UP000000803 UP000001292 UP000000304 UP000008711 UP000192221 UP000094527 UP000076858 UP000001819 UP000008744 UP000075884 UP000092553 UP000007798
UP000092461 UP000245037 UP000235965 UP000007266 UP000076408 UP000075882 UP000075886 UP000075901 UP000069272 UP000002320 UP000075900 UP000069940 UP000249989 UP000030765 UP000008792 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000075920 UP000008820 UP000000673 UP000091820 UP000007819 UP000027135 UP000095300 UP000075885 UP000092443 UP000009192 UP000078200 UP000075880 UP000095301 UP000002282 UP000000803 UP000001292 UP000000304 UP000008711 UP000192221 UP000094527 UP000076858 UP000001819 UP000008744 UP000075884 UP000092553 UP000007798
Pfam
PF04716 ETC_C1_NDUFA5
Interpro
IPR006806
NDUFA5
ProteinModelPortal
H9J225
Q1G0N4
A0A0L7KSC1
A0A212FEZ8
A0A0N1IIK2
S4NXE9
+ More
I4DK45 A0A2W1BUY1 A0A2A4K3L6 A0A1E1WDB2 A0A2H1V0C5 A0A1B0CRS4 A0A1B0CJQ6 A0A2P8XLU3 A0A2J7R2Q5 A0A1L8E0G1 A0A1Y1MUQ6 D6X292 A0A182Y179 A0A182LJ73 A0A182QLJ1 V5GFE4 A0A182SZT9 A0A0K8TSU9 Q1HQT0 A0A2M4CNF4 A0A1Q3FKX8 A0A182FDM6 B0WLS3 A0A023EFW7 A0A2H1WMG4 A0A2M3Z686 A0A182RRV3 A0A182GZH1 A0A084VCA6 B4LB56 A0A182VB56 A0A182UDI7 A0A182XMZ3 Q7PNP3 A0A182HWU9 A0A2M4C509 A0A2M4C4M6 T1DJH3 A0A182WB26 A0A1S4F8R1 A0A2M4C3B0 Q0IFI1 C4WUD5 W5JFR1 A0A1A9WTA2 A0A2S2P7F5 J9K5T5 A0A2M4AFK0 A0A067RC26 A0A2M4AFZ0 A0A1I8QB73 A0A182PHK0 A0A1A9Y053 B4L9G6 A0A1A9UG23 A0A182JBP1 A0A1I8M7P5 A0A2H8TJC9 B4PEG9 Q9VTB4 B4HDR3 B4QP91 B3NCN0 A0A0A1WEU3 A0A1W4UV17 A0A1L8EEP4 A0A1D2MW47 A0A0P5F2H7 A0A0P4XNG0 A0A0P5AFP3 Q29E06 B4H439 A0A0P5HUK1 A0A0P5YA62 A0A0N8D3P7 A0A0P5HQ27 A0A0N8DB15 A0A0P6D5H1 A0A0P6D7H0 A0A0P6H4W8 A0A0P5D8A8 A0A293M0Q4 A0A1L8EF84 A0A182NGT9 A0A1E1X0T2 W8C0Z7 A0A0M3QW54 A0A0P5TXP9 A0A034WVB1 B4MLV5 A0A0N8DM33 A0A1B6KJS4 A0A224YWE1 A0A224Z4Q8
I4DK45 A0A2W1BUY1 A0A2A4K3L6 A0A1E1WDB2 A0A2H1V0C5 A0A1B0CRS4 A0A1B0CJQ6 A0A2P8XLU3 A0A2J7R2Q5 A0A1L8E0G1 A0A1Y1MUQ6 D6X292 A0A182Y179 A0A182LJ73 A0A182QLJ1 V5GFE4 A0A182SZT9 A0A0K8TSU9 Q1HQT0 A0A2M4CNF4 A0A1Q3FKX8 A0A182FDM6 B0WLS3 A0A023EFW7 A0A2H1WMG4 A0A2M3Z686 A0A182RRV3 A0A182GZH1 A0A084VCA6 B4LB56 A0A182VB56 A0A182UDI7 A0A182XMZ3 Q7PNP3 A0A182HWU9 A0A2M4C509 A0A2M4C4M6 T1DJH3 A0A182WB26 A0A1S4F8R1 A0A2M4C3B0 Q0IFI1 C4WUD5 W5JFR1 A0A1A9WTA2 A0A2S2P7F5 J9K5T5 A0A2M4AFK0 A0A067RC26 A0A2M4AFZ0 A0A1I8QB73 A0A182PHK0 A0A1A9Y053 B4L9G6 A0A1A9UG23 A0A182JBP1 A0A1I8M7P5 A0A2H8TJC9 B4PEG9 Q9VTB4 B4HDR3 B4QP91 B3NCN0 A0A0A1WEU3 A0A1W4UV17 A0A1L8EEP4 A0A1D2MW47 A0A0P5F2H7 A0A0P4XNG0 A0A0P5AFP3 Q29E06 B4H439 A0A0P5HUK1 A0A0P5YA62 A0A0N8D3P7 A0A0P5HQ27 A0A0N8DB15 A0A0P6D5H1 A0A0P6D7H0 A0A0P6H4W8 A0A0P5D8A8 A0A293M0Q4 A0A1L8EF84 A0A182NGT9 A0A1E1X0T2 W8C0Z7 A0A0M3QW54 A0A0P5TXP9 A0A034WVB1 B4MLV5 A0A0N8DM33 A0A1B6KJS4 A0A224YWE1 A0A224Z4Q8
PDB
5GUP
E-value=1.14428e-22,
Score=256
Ontologies
PATHWAY
GO
PANTHER
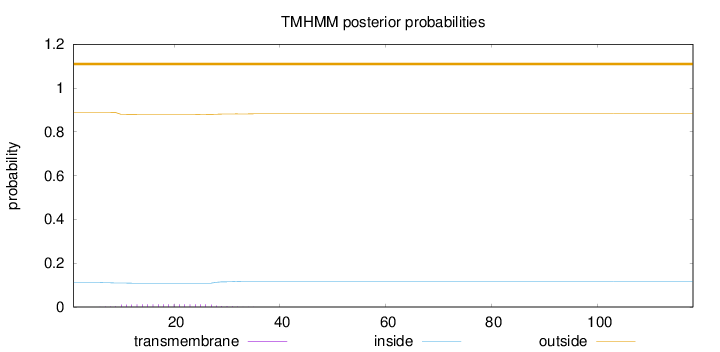
Topology
Length:
118
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23627
Exp number, first 60 AAs:
0.23627
Total prob of N-in:
0.11174
outside
1 - 118
Population Genetic Test Statistics
Pi
147.925901
Theta
165.109058
Tajima's D
-0.304063
CLR
87.262546
CSRT
0.290835458227089
Interpretation
Uncertain
