Gene
KWMTBOMO02940
Pre Gene Modal
BGIBMGA003633
Annotation
E3_ubiquitin-protein_ligase_Topors_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.809
Sequence
CDS
ATGGAGGCAGTGCCACAATCTGTTGATGATGTCTCTGATGCCAACTCTAGCAACATAAAAAATGACAACAGCAGTCAACACCATGAATCCAATAGTAGGGAATCACCTCCTCCTAACTGTGCAATATGTCTAGGAATGTGTCAAAACAAATCATTTACAGACTCTTGTTTGCACCAGTTTTGTTTTAAGTGTCTTGTAACATGGAGTAAGGTAAAAGCAGAATGCCCTTTATGCAAACAAAGCTTCAAAATAATAAAATACAATGTGATCAGCAGTTGTCAATTTGAAGAGTACAAAGTGGAAGAGCGCCAAAATGAAACAGTCGAAAGTGTTGTTATCGAAGCCCTGACAAGTATCTTGTTCAGTTACAGAACAATTCTGACATCTCAACACAATTCATTGGCCGTTGAAGACTTCTTGCTGCAACACTACCCACAGTCCTCAAACGCGTTAAGCCTGCCATCCCGCAGCAGATCTTCAGCCTCTTTTCGTCGCATCATCTACGAACGTAACTTACGGGCGCAACCTCTCCCCGATCACACCGGCCGCTTTCGAAATTGTAGTCCCGGATACTACAGGTACAATATGTTTGAAATATATCGCCTCGTTCCCTGGTTAAACAGGGAACTCAACTACTTACTGAATGAAAACGTCAGCCACATTTCTTACGTATTAATGCGAATTTTGGAAAGATTACTCCTCTATGACATAACTTCATCACAGTTTCGAGAAGCGATGCGACGCTTTTTCGGCGAGAGAACGGAACATTTCTTGCATGAGTTACACTGCTTTGCTACCTCTCCATATGATATGAATGGTTATGATCGTAATGTGCAATACACCACCGAATCGCGCGCGTTGACTATGGTCAATGAGGTAATATCTACTTCAGAATCGGACGGGAGCATTGACACCGACGTCGCTATGATACCGAATTCTGTGGCCGCTGAGCCTCCCGCCGGCCCTTCGGGGGTGCGACGAGCCCCGCCAAGAGCTGCTCTCGCAGCACATGCCGTCATGCCTATCGAAACCATCGCCAATTCCGACACCGACGACTCTTCAGAAGTGATGGTTGTTGGTTACATCAAGCCCCCGAAAGATCGAACACCGGAGCTGGTCGACCTGGTCGGCTCAGATAGTGATGTAGTTGTTGAGGAGAGTTCACGGGCCAAGTCTGGGCCGTCGGATACGAGACCGACGATCCCGCCTGTAAGTAAGCTCAAATTGAAGAGGAGCCGCGCTGGTGGGACGGAGTTGTCCGATTCGGATACGGACGACTCGTATCAATCACCAGCCCGTCGTCGTCGCCGACCTCGCATCTCGAACTCTGGTATGATGGCAAGCACGCATCGTACTACGCCAGAACCTACCTCCACCGCCTCAGAATACGACACTGCAGATGAGTTGACCCCTCCAAGTTAA
Protein
MEAVPQSVDDVSDANSSNIKNDNSSQHHESNSRESPPPNCAICLGMCQNKSFTDSCLHQFCFKCLVTWSKVKAECPLCKQSFKIIKYNVISSCQFEEYKVEERQNETVESVVIEALTSILFSYRTILTSQHNSLAVEDFLLQHYPQSSNALSLPSRSRSSASFRRIIYERNLRAQPLPDHTGRFRNCSPGYYRYNMFEIYRLVPWLNRELNYLLNENVSHISYVLMRILERLLLYDITSSQFREAMRRFFGERTEHFLHELHCFATSPYDMNGYDRNVQYTTESRALTMVNEVISTSESDGSIDTDVAMIPNSVAAEPPAGPSGVRRAPPRAALAAHAVMPIETIANSDTDDSSEVMVVGYIKPPKDRTPELVDLVGSDSDVVVEESSRAKSGPSDTRPTIPPVSKLKLKRSRAGGTELSDSDTDDSYQSPARRRRRPRISNSGMMASTHRTTPEPTSTASEYDTADELTPPS
Summary
Uniprot
A0A194PQ17
S4PXJ8
A0A2H1V0F9
A0A2A4K3K9
A0A2A4K488
A0A2W1BKF3
+ More
A0A212FF06 A0A0L7KSU0 A0A232F510 A0A310S9I5 A0A0N0U7H3 E0VQ22 A0A088AMA8 K7J3U1 A0A2A3E8K0 A0A195C1T3 A0A1Z5LC42 A0A195DAZ3 A0A151K0G1 A0A2J7R2N0 A0A0L7RC51 F4WUF1 A0A151X7V5 A0A026W8T2 A0A0K2V0G4 A0A293LJB5 A0A2R5LI95 E2ASD1 A0A0C9QRK3 A0A0C9RHC3 A0A0N8C3G5 U4UG34 A0A023GFN8 A0A023EZ23 A0A224XJQ6
A0A212FF06 A0A0L7KSU0 A0A232F510 A0A310S9I5 A0A0N0U7H3 E0VQ22 A0A088AMA8 K7J3U1 A0A2A3E8K0 A0A195C1T3 A0A1Z5LC42 A0A195DAZ3 A0A151K0G1 A0A2J7R2N0 A0A0L7RC51 F4WUF1 A0A151X7V5 A0A026W8T2 A0A0K2V0G4 A0A293LJB5 A0A2R5LI95 E2ASD1 A0A0C9QRK3 A0A0C9RHC3 A0A0N8C3G5 U4UG34 A0A023GFN8 A0A023EZ23 A0A224XJQ6
Pubmed
EMBL
KQ459597
KPI95058.1
GAIX01004913
JAA87647.1
ODYU01000103
SOQ34299.1
+ More
NWSH01000167 PCG78845.1 PCG78846.1 KZ149992 PZC75562.1 AGBW02008872 OWR52322.1 JTDY01006277 KOB66150.1 NNAY01000925 OXU25871.1 KQ762882 OAD55320.1 KQ435711 KOX79551.1 DS235389 EEB15478.1 KZ288340 PBC27784.1 KQ978344 KYM94807.1 GFJQ02002013 JAW04957.1 KQ981042 KYN10058.1 KQ981253 KYN44195.1 NEVH01007828 PNF35083.1 KQ414615 KOC68567.1 GL888359 EGI62177.1 KQ982431 KYQ56457.1 KK107335 QOIP01000013 EZA52465.1 RLU15183.1 HACA01026637 CDW43998.1 GFWV01004083 MAA28813.1 GGLE01005116 MBY09242.1 GL442284 EFN63665.1 GBYB01006324 GBYB01006325 GBYB01006327 JAG76091.1 JAG76092.1 JAG76094.1 GBYB01006326 JAG76093.1 GDIQ01118539 JAL33187.1 KB632173 ERL89566.1 GBBM01003635 JAC31783.1 GBBI01004731 JAC13981.1 GFTR01007604 JAW08822.1
NWSH01000167 PCG78845.1 PCG78846.1 KZ149992 PZC75562.1 AGBW02008872 OWR52322.1 JTDY01006277 KOB66150.1 NNAY01000925 OXU25871.1 KQ762882 OAD55320.1 KQ435711 KOX79551.1 DS235389 EEB15478.1 KZ288340 PBC27784.1 KQ978344 KYM94807.1 GFJQ02002013 JAW04957.1 KQ981042 KYN10058.1 KQ981253 KYN44195.1 NEVH01007828 PNF35083.1 KQ414615 KOC68567.1 GL888359 EGI62177.1 KQ982431 KYQ56457.1 KK107335 QOIP01000013 EZA52465.1 RLU15183.1 HACA01026637 CDW43998.1 GFWV01004083 MAA28813.1 GGLE01005116 MBY09242.1 GL442284 EFN63665.1 GBYB01006324 GBYB01006325 GBYB01006327 JAG76091.1 JAG76092.1 JAG76094.1 GBYB01006326 JAG76093.1 GDIQ01118539 JAL33187.1 KB632173 ERL89566.1 GBBM01003635 JAC31783.1 GBBI01004731 JAC13981.1 GFTR01007604 JAW08822.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194PQ17
S4PXJ8
A0A2H1V0F9
A0A2A4K3K9
A0A2A4K488
A0A2W1BKF3
+ More
A0A212FF06 A0A0L7KSU0 A0A232F510 A0A310S9I5 A0A0N0U7H3 E0VQ22 A0A088AMA8 K7J3U1 A0A2A3E8K0 A0A195C1T3 A0A1Z5LC42 A0A195DAZ3 A0A151K0G1 A0A2J7R2N0 A0A0L7RC51 F4WUF1 A0A151X7V5 A0A026W8T2 A0A0K2V0G4 A0A293LJB5 A0A2R5LI95 E2ASD1 A0A0C9QRK3 A0A0C9RHC3 A0A0N8C3G5 U4UG34 A0A023GFN8 A0A023EZ23 A0A224XJQ6
A0A212FF06 A0A0L7KSU0 A0A232F510 A0A310S9I5 A0A0N0U7H3 E0VQ22 A0A088AMA8 K7J3U1 A0A2A3E8K0 A0A195C1T3 A0A1Z5LC42 A0A195DAZ3 A0A151K0G1 A0A2J7R2N0 A0A0L7RC51 F4WUF1 A0A151X7V5 A0A026W8T2 A0A0K2V0G4 A0A293LJB5 A0A2R5LI95 E2ASD1 A0A0C9QRK3 A0A0C9RHC3 A0A0N8C3G5 U4UG34 A0A023GFN8 A0A023EZ23 A0A224XJQ6
PDB
1CHC
E-value=0.000143992,
Score=108
Ontologies
GO
Topology
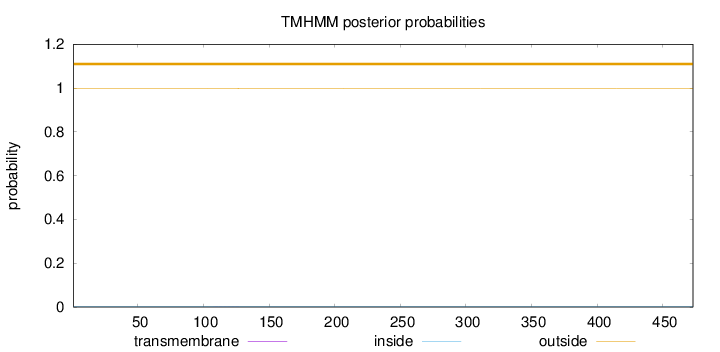
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00448
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00246
outside
1 - 473
Population Genetic Test Statistics
Pi
275.644617
Theta
210.18534
Tajima's D
0.937793
CLR
0.211143
CSRT
0.646167691615419
Interpretation
Uncertain
