Gene
KWMTBOMO02939
Pre Gene Modal
BGIBMGA003634
Annotation
E3_ubiquitin-protein_ligase_Topors_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 4.704
Sequence
CDS
ATGGCGACTATACCACAATCTTTGGATGATATCTCTGAAGCAAACCCAAGCAGCATAAAGAGTGATGATAGCAGTCCACATCATGACAACAGCAGAGGGTCGCCACCTCCTAACTGTGCAATATGTCTAGGAACATGTAAAAACAAATCATTCACTGATTCTTGCTTGCACCAATTTTGTTTCAACTGTCTTGTAACTTGGAGTAAGGTGAAAGCAGAATGTCCATTATGCAAGCAAAATTTTAAATCAATAATTTATAATGTTCGTGCCAATCAACAATATGAAGAATACCAAGTGGAACAGCGCCAAGTTGAATCTAGCCAGAGAATTGATATTGATACACTCACAAACAGAAGGTTTAGATACAGAACTACATTGACATTGCCACGACGGCAGTCGATAGCCTTCCAGCAGTTGTTGTTACGGCACTACCCACTTGTGGCAAATGTGTTGAGTCCACCACGTCGTCGACGCTCTCCGGCTTCCTTCCGTCGCACTGTGTACCGACACAACCTGTGGGCGCGACCACTCCCCGACTTCACTGGACGGTTTCGTGACTGTACGCCAGAATTCTATAGATCTAATGCCACACAGTTACATCGTCTCGTGCCCTGGTTGAATAGGGAGCTTAGCTACTTATTAAATGGAAACGTAGGTCATATTTCTTACGTGCTGACCAGAATCTTGGAAATGTTGCCTCACCATCACATAACTTTCGGGACCGAGTTTCGGGAAGCGATGACACGTTTCTTTGGTGACAGGACAGACCATTTCTTACACGAGTTGTACTGCTTCGCGTCAACACCCTACGACATGACCGGATATGATCGTAATGTGCAATACACGACTGATTCGCGTATGTCGACAATGGTCAACGAAGTGATATCTAGCTCGGAGTCAGATGGAAGTGTCGACTCGGATATCGTGATGGTGTCGAGTTCATCTGCCGCCGAACCTTCCGCCGGTCCCTCGGGAGTACCGCGAGCTCCGCCCCGGGCCGCCCTCGCAGCCAACAATATCATTCCCATTGAAACAATTTCACATTCTGACACTGACGACGACTCATCCGAAGTAATGGTCGTCGGCTATATTAAACCCCCTCAGGATCGAACTCCAGAACTCGTTGATCTGGTCGGATCTGATAGCGACGTAGTGATCGCCGAAACCACGCAACCTCAACCTGGACCGTCCGACACGAATGCCGGCCCTTTTGTGAAGCTTACTTTGAAAAGAGACCGTCCTGAAGGGTCGAATTGGTCTGGTTCGGACTCTGACGACTCGTACCGTCCGCTAAGGCGTCGTCGCCGGCGGCCGCGCTCCTCCGACTCCGCCACGACGGCAGACACGCGCCGCACCACGCCCGAACCTGCCTCCTCCACCTCCGCTTCGTCCGACGCCGATCAGTCGTCAACAGACACGTCACTATCTGACGACAGAAAAACCGGTGCCACCAGAAAGAAGATCAGAAAGAGAAGCAGCAGGGAAGGCGCAACATTTCGCCGGGAAAGAAATGGAAGTGGAAGACGTTCGTCGCCCGGTCAACGACGCGATAACAAAGTAAATACCGACTCGAGTCATCCGGATCGAGACCGACGAAGTAAAAAGAAGTCACATTCAGGAAAAGGAGTGAAATGTTCTAGACGTCTCGAGGAGCCTCCCCCAGAAGCTCGGCCGACGGAATCCGCGGGACGGAAGTTGAGGAGCGTCGTCACCGTCATGCGCGGCCGGACCGCCGGGCCCGACAGGAGGTCCAGCATCTCTGACTCAGACGACGACCTGCCTCTTAATCTCACTGTACAGAAAAAGCAACCTCCCTCCTAG
Protein
MATIPQSLDDISEANPSSIKSDDSSPHHDNSRGSPPPNCAICLGTCKNKSFTDSCLHQFCFNCLVTWSKVKAECPLCKQNFKSIIYNVRANQQYEEYQVEQRQVESSQRIDIDTLTNRRFRYRTTLTLPRRQSIAFQQLLLRHYPLVANVLSPPRRRRSPASFRRTVYRHNLWARPLPDFTGRFRDCTPEFYRSNATQLHRLVPWLNRELSYLLNGNVGHISYVLTRILEMLPHHHITFGTEFREAMTRFFGDRTDHFLHELYCFASTPYDMTGYDRNVQYTTDSRMSTMVNEVISSSESDGSVDSDIVMVSSSSAAEPSAGPSGVPRAPPRAALAANNIIPIETISHSDTDDDSSEVMVVGYIKPPQDRTPELVDLVGSDSDVVIAETTQPQPGPSDTNAGPFVKLTLKRDRPEGSNWSGSDSDDSYRPLRRRRRRPRSSDSATTADTRRTTPEPASSTSASSDADQSSTDTSLSDDRKTGATRKKIRKRSSREGATFRRERNGSGRRSSPGQRRDNKVNTDSSHPDRDRRSKKKSHSGKGVKCSRRLEEPPPEARPTESAGRKLRSVVTVMRGRTAGPDRRSSISDSDDDLPLNLTVQKKQPPS
Summary
Uniprot
Pubmed
Proteomes
Gene 3D
ProteinModelPortal
PDB
1CHC
E-value=1.56962e-05,
Score=118
Ontologies
GO
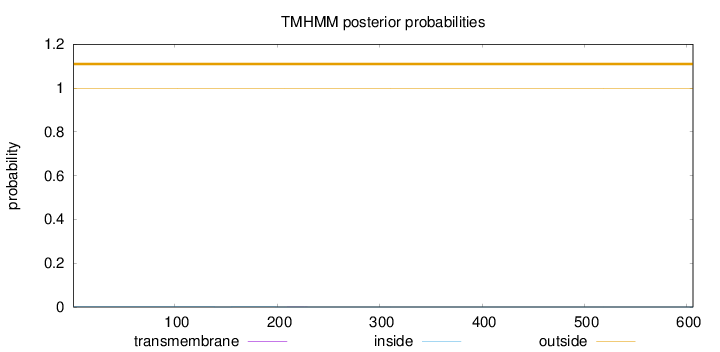
Topology
Length:
606
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00423
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00059
outside
1 - 606
Population Genetic Test Statistics
Pi
206.171878
Theta
190.87395
Tajima's D
0.047765
CLR
0.028208
CSRT
0.380680965951702
Interpretation
Uncertain
