Gene
KWMTBOMO02938 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003561
Annotation
p38_map_kinase_[Bombyx_mori]
Full name
Mitogen-activated protein kinase
Location in the cell
Cytoplasmic Reliability : 1.89 Mitochondrial Reliability : 1.657
Sequence
CDS
ATGCCTCGTTTCCATAAAGTAGAAATTAATAAAACTGAATGGATAGTCCCGGAGCGATATCAGATGCTTACACCTGTAGGTTCCGGCGCTTACGGTCAGGTTTGTTCTGCAATAGATGCCCAACATAGTATGAAAGTGGCTATTAAAAAGTTAGCCAGACCTTTTCAATCAGCTGTTCATGCAAAGAGAACATACAGAGAGTTGCGAATGTTGAAACATATGAACCATGAGAATGTTATTGGATTGCTGGATGTGTTTACACCGGAGAAAACATTAGAAGATTTTCAGCAAGTATACTTAGTGACCCATCTAATGGGTGCTGATCTGAATAACATCGTACGGACACAGAAGCTTTCTGATGACCACGTTCAGTTTCTGGTATACCAGATTTTACGTGGACTCAAATACATACATTCGGCTGGCATCATTCATAGGGATTTAAAACCCTCTAACATAGCTGTGAATGAGGACTGTGAGTTAAAAATCTTAGATTTTGGCTTGGCGAGACCCACTGAGACTGAAATGACAGGTTATGTGGCAACAAGATGGTATCGTGCGCCAGAGATAATGCTCAATTGGATGCATTATAACCAAACTGTGGACATTTGGTCTGTGGGTTGTATTATGGCTGAGTTGCTGACAGGAAGAACTTTGTTCCCCGGTACTGACCATATTCATCAATTGAATTTAATCATGGAGATACTCGGCACGCCCGCTCAGGAGTTTATGCAGAAAATATCGTCCGAGTCTGCACGTAACTACATCCAGTCTCTGCCGGCTTTGAAGAGACGCGACTTCCGCGAGGTGTTCCGCGGCGCGAACCCACTCGCCATCAACCTGTTGGAACTGATGCTCGAACTGGATGCCGATAAACGCATAACGGCAGAACAAGCGCTCGCGCACGAGTACTTGGCCCAGTACGCGGACCCGACCGACGAGCCGGTTTCTGCGCCCTACGACCAGAGCTTCGAAGACATGGAGCTGCCTGTAGACAAGTGGAAAGGTGATTTTTTTTATTGCACATATAGGTGGACGAGCTCTCACAGCCCACCTGGTGTAAATGCCGCCACCCACCTTGAGATATGA
Protein
MPRFHKVEINKTEWIVPERYQMLTPVGSGAYGQVCSAIDAQHSMKVAIKKLARPFQSAVHAKRTYRELRMLKHMNHENVIGLLDVFTPEKTLEDFQQVYLVTHLMGADLNNIVRTQKLSDDHVQFLVYQILRGLKYIHSAGIIHRDLKPSNIAVNEDCELKILDFGLARPTETEMTGYVATRWYRAPEIMLNWMHYNQTVDIWSVGCIMAELLTGRTLFPGTDHIHQLNLIMEILGTPAQEFMQKISSESARNYIQSLPALKRRDFREVFRGANPLAINLLELMLELDADKRITAEQALAHEYLAQYADPTDEPVSAPYDQSFEDMELPVDKWKGDFFYCTYRWTSSHSPPGVNAATHLEI
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Uniprot
Q3C2E3
A0A2A4K4K7
A0A212FF41
A0A023JNR8
A0A1B0YTK4
A0A0N1PHN0
+ More
A0A2J7QC17 A0A2Z5U1Y0 A0A310SHK9 E2B3L1 A0A154P726 F8QQF2 F4X361 E2AIS5 E0VR05 A0A158NPL5 A0A1B6CXK1 A0A0C9QVW1 A0A195FFU9 A0A067RWU2 A0A0J7NCT8 A0A087ZYD6 A0A023EQK1 A0A1Q3FXJ6 Q1L0R1 A0A2J7QC09 U5EY70 A0A151IDR9 A0A1B6FMW4 A0A2H1V829 A0A182JIC7 A0A026WTT7 A0A084WM35 A0A195BEV0 A0A151IVK5 A0A151WEG4 A0A182X017 A0A182US54 A0A182KT27 A0A182HPR1 A0A336M6U7 A0A2H8TSD8 A0A2S2QW05 A0A1B6KES4 A0A182QL11 J9KAX7 A0A2A3E4W9 A0A2M3ZFN3 A0A2M4A467 A0A182FVA0 A0A2M4BS21 G3F9Y2 A0A2M4CTJ6 A0A0K8TPY2 A0A3L8DZU2 A0A1B6EAR8 T1E3S8 A0A165A8C5 A0A0P6IIL7 A0A0M8ZU21 A0A069DTG8 A0A023FAE4 A0A0P6BVV9 E9FQM3 A0A0P5VMX5 A0A1L8DKN4 A0A0P5VMX8 A0A0P4VP85 R4G4C2 A0A1W4WYL0 A0A0V0GAP6 A0A182YC55 A0A1Y1NHD4 Q16YY2 A0A182GRN6 A0A1J1IIP5 L7X0J9 A0A0P5ZL25 A0A0A7D769 A0A2S2NND6 A0A0P4WBX5 A0A023JBY8 A0A0P5SFK8 A0A1B6EUA3 A0A0P5HV78 A0A0P5SSW0 A0A146LVE3 A0A1A9W850 A0A221I025 A0A0P5SPW4 A0A0L0C6N3 A0A0P5DZD2 A0A288ZBA2 A7BHS5 A0A1I8NA24 A0A1L8EC74 A0A1B0A5N9 A0A1B0GB82 A0A2P2I1S0 A0A1B6MB35
A0A2J7QC17 A0A2Z5U1Y0 A0A310SHK9 E2B3L1 A0A154P726 F8QQF2 F4X361 E2AIS5 E0VR05 A0A158NPL5 A0A1B6CXK1 A0A0C9QVW1 A0A195FFU9 A0A067RWU2 A0A0J7NCT8 A0A087ZYD6 A0A023EQK1 A0A1Q3FXJ6 Q1L0R1 A0A2J7QC09 U5EY70 A0A151IDR9 A0A1B6FMW4 A0A2H1V829 A0A182JIC7 A0A026WTT7 A0A084WM35 A0A195BEV0 A0A151IVK5 A0A151WEG4 A0A182X017 A0A182US54 A0A182KT27 A0A182HPR1 A0A336M6U7 A0A2H8TSD8 A0A2S2QW05 A0A1B6KES4 A0A182QL11 J9KAX7 A0A2A3E4W9 A0A2M3ZFN3 A0A2M4A467 A0A182FVA0 A0A2M4BS21 G3F9Y2 A0A2M4CTJ6 A0A0K8TPY2 A0A3L8DZU2 A0A1B6EAR8 T1E3S8 A0A165A8C5 A0A0P6IIL7 A0A0M8ZU21 A0A069DTG8 A0A023FAE4 A0A0P6BVV9 E9FQM3 A0A0P5VMX5 A0A1L8DKN4 A0A0P5VMX8 A0A0P4VP85 R4G4C2 A0A1W4WYL0 A0A0V0GAP6 A0A182YC55 A0A1Y1NHD4 Q16YY2 A0A182GRN6 A0A1J1IIP5 L7X0J9 A0A0P5ZL25 A0A0A7D769 A0A2S2NND6 A0A0P4WBX5 A0A023JBY8 A0A0P5SFK8 A0A1B6EUA3 A0A0P5HV78 A0A0P5SSW0 A0A146LVE3 A0A1A9W850 A0A221I025 A0A0P5SPW4 A0A0L0C6N3 A0A0P5DZD2 A0A288ZBA2 A7BHS5 A0A1I8NA24 A0A1L8EC74 A0A1B0A5N9 A0A1B0GB82 A0A2P2I1S0 A0A1B6MB35
EC Number
2.7.11.24
Pubmed
EMBL
AB208585
BAE46743.1
NWSH01000167
PCG78844.1
AGBW02008872
OWR52323.1
+ More
KC895497 KZ149992 AHL46461.1 PZC75561.1 KU358549 ANO46359.1 KQ460226 KPJ16463.1 NEVH01016292 PNF26119.1 FX985764 BBA93651.1 KQ769179 OAD52922.1 GL445346 EFN89763.1 KQ434829 KZC07746.1 GU321334 GU321336 ADT91685.1 GL888609 EGI59042.1 GL439874 EFN66664.1 DS235442 EEB15811.1 ADTU01000325 GEDC01019098 JAS18200.1 GBYB01004767 JAG74534.1 KQ981606 KYN39575.1 KK852415 KDR24374.1 LBMM01006692 KMQ90390.1 GAPW01002233 JAC11365.1 GFDL01002741 JAV32304.1 DQ270546 CH477506 ABB82553.1 EAT39844.1 PNF26120.1 GANO01002169 JAB57702.1 KQ977978 KYM98399.1 GECZ01026958 GECZ01018223 GECZ01013569 GECZ01011792 GECZ01010640 GECZ01010296 GECZ01007977 GECZ01005023 JAS42811.1 JAS51546.1 JAS56200.1 JAS57977.1 JAS59129.1 JAS59473.1 JAS61792.1 JAS64746.1 ODYU01001155 SOQ36967.1 KK107106 EZA59427.1 ATLV01024368 ATLV01024369 ATLV01024370 ATLV01024371 ATLV01024372 ATLV01024373 KE525351 KFB51279.1 KQ976509 KYM82690.1 KQ980895 KYN11740.1 KQ983238 KYQ46244.1 APCN01003267 UFQT01000641 SSX26024.1 GFXV01004313 MBW16118.1 GGMS01012640 MBY81843.1 GEBQ01032215 GEBQ01030031 JAT07762.1 JAT09946.1 AXCN02001696 ABLF02021646 KZ288369 PBC26777.1 GGFM01006568 MBW27319.1 GGFK01002117 MBW35438.1 GGFJ01006735 MBW55876.1 JF505508 AEA92685.1 GGFL01004421 MBW68599.1 GDAI01001648 JAI15955.1 QOIP01000002 RLU25822.1 GEDC01002283 JAS35015.1 GALA01000100 JAA94752.1 LRGB01000642 KZS17293.1 GDIQ01021347 JAN73390.1 KQ435847 KOX71145.1 GBGD01001927 JAC86962.1 GBBI01000743 JAC17969.1 GDIP01008892 JAM94823.1 GL732523 EFX90341.1 GDIP01097858 JAM05857.1 GFDF01007157 JAV06927.1 GDIP01097859 JAM05856.1 GDKW01001783 JAI54812.1 ACPB03000101 GAHY01001495 JAA76015.1 GECL01001645 JAP04479.1 GEZM01006949 JAV95536.1 EAT39843.1 JXUM01082844 JXUM01082845 JXUM01082846 JXUM01082847 JXUM01082848 JXUM01082849 JXUM01082850 JXUM01082851 JXUM01082852 JXUM01082853 KQ563340 KXJ74011.1 CVRI01000051 CRK99642.1 KC012937 AGC92010.1 GDIP01042456 JAM61259.1 JX489772 KF582665 AGK89796.1 AII32447.1 GGMR01006082 MBY18701.1 GDRN01054823 JAI66059.1 KC711047 MH341515 AHH29322.1 QBA57530.1 GDIP01187247 GDIP01140357 JAL63357.1 GECZ01028237 JAS41532.1 GDIQ01221751 JAK29974.1 GDIQ01227819 GDIP01135756 GDIP01134405 JAK23906.1 JAL67958.1 GDHC01007460 JAQ11169.1 KY765908 ASM46958.1 GDIP01187246 GDIP01140356 JAL63358.1 JRES01000824 KNC28078.1 GDIP01149595 GDIQ01218069 GDIP01075755 JAJ73807.1 JAK33656.1 KX893515 ASU54245.1 AB277828 BAF75366.1 GFDG01002580 JAV16219.1 CCAG010011560 IACF01002270 LAB67931.1 GEBQ01006837 GEBQ01002711 JAT33140.1 JAT37266.1
KC895497 KZ149992 AHL46461.1 PZC75561.1 KU358549 ANO46359.1 KQ460226 KPJ16463.1 NEVH01016292 PNF26119.1 FX985764 BBA93651.1 KQ769179 OAD52922.1 GL445346 EFN89763.1 KQ434829 KZC07746.1 GU321334 GU321336 ADT91685.1 GL888609 EGI59042.1 GL439874 EFN66664.1 DS235442 EEB15811.1 ADTU01000325 GEDC01019098 JAS18200.1 GBYB01004767 JAG74534.1 KQ981606 KYN39575.1 KK852415 KDR24374.1 LBMM01006692 KMQ90390.1 GAPW01002233 JAC11365.1 GFDL01002741 JAV32304.1 DQ270546 CH477506 ABB82553.1 EAT39844.1 PNF26120.1 GANO01002169 JAB57702.1 KQ977978 KYM98399.1 GECZ01026958 GECZ01018223 GECZ01013569 GECZ01011792 GECZ01010640 GECZ01010296 GECZ01007977 GECZ01005023 JAS42811.1 JAS51546.1 JAS56200.1 JAS57977.1 JAS59129.1 JAS59473.1 JAS61792.1 JAS64746.1 ODYU01001155 SOQ36967.1 KK107106 EZA59427.1 ATLV01024368 ATLV01024369 ATLV01024370 ATLV01024371 ATLV01024372 ATLV01024373 KE525351 KFB51279.1 KQ976509 KYM82690.1 KQ980895 KYN11740.1 KQ983238 KYQ46244.1 APCN01003267 UFQT01000641 SSX26024.1 GFXV01004313 MBW16118.1 GGMS01012640 MBY81843.1 GEBQ01032215 GEBQ01030031 JAT07762.1 JAT09946.1 AXCN02001696 ABLF02021646 KZ288369 PBC26777.1 GGFM01006568 MBW27319.1 GGFK01002117 MBW35438.1 GGFJ01006735 MBW55876.1 JF505508 AEA92685.1 GGFL01004421 MBW68599.1 GDAI01001648 JAI15955.1 QOIP01000002 RLU25822.1 GEDC01002283 JAS35015.1 GALA01000100 JAA94752.1 LRGB01000642 KZS17293.1 GDIQ01021347 JAN73390.1 KQ435847 KOX71145.1 GBGD01001927 JAC86962.1 GBBI01000743 JAC17969.1 GDIP01008892 JAM94823.1 GL732523 EFX90341.1 GDIP01097858 JAM05857.1 GFDF01007157 JAV06927.1 GDIP01097859 JAM05856.1 GDKW01001783 JAI54812.1 ACPB03000101 GAHY01001495 JAA76015.1 GECL01001645 JAP04479.1 GEZM01006949 JAV95536.1 EAT39843.1 JXUM01082844 JXUM01082845 JXUM01082846 JXUM01082847 JXUM01082848 JXUM01082849 JXUM01082850 JXUM01082851 JXUM01082852 JXUM01082853 KQ563340 KXJ74011.1 CVRI01000051 CRK99642.1 KC012937 AGC92010.1 GDIP01042456 JAM61259.1 JX489772 KF582665 AGK89796.1 AII32447.1 GGMR01006082 MBY18701.1 GDRN01054823 JAI66059.1 KC711047 MH341515 AHH29322.1 QBA57530.1 GDIP01187247 GDIP01140357 JAL63357.1 GECZ01028237 JAS41532.1 GDIQ01221751 JAK29974.1 GDIQ01227819 GDIP01135756 GDIP01134405 JAK23906.1 JAL67958.1 GDHC01007460 JAQ11169.1 KY765908 ASM46958.1 GDIP01187246 GDIP01140356 JAL63358.1 JRES01000824 KNC28078.1 GDIP01149595 GDIQ01218069 GDIP01075755 JAJ73807.1 JAK33656.1 KX893515 ASU54245.1 AB277828 BAF75366.1 GFDG01002580 JAV16219.1 CCAG010011560 IACF01002270 LAB67931.1 GEBQ01006837 GEBQ01002711 JAT33140.1 JAT37266.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000235965
UP000008237
UP000076502
+ More
UP000007755 UP000000311 UP000009046 UP000005205 UP000078541 UP000027135 UP000036403 UP000005203 UP000008820 UP000078542 UP000075880 UP000053097 UP000030765 UP000078540 UP000078492 UP000075809 UP000076407 UP000075903 UP000075882 UP000075840 UP000075886 UP000007819 UP000242457 UP000069272 UP000279307 UP000076858 UP000053105 UP000000305 UP000015103 UP000192223 UP000076408 UP000069940 UP000249989 UP000183832 UP000091820 UP000037069 UP000095301 UP000092445 UP000092444
UP000007755 UP000000311 UP000009046 UP000005205 UP000078541 UP000027135 UP000036403 UP000005203 UP000008820 UP000078542 UP000075880 UP000053097 UP000030765 UP000078540 UP000078492 UP000075809 UP000076407 UP000075903 UP000075882 UP000075840 UP000075886 UP000007819 UP000242457 UP000069272 UP000279307 UP000076858 UP000053105 UP000000305 UP000015103 UP000192223 UP000076408 UP000069940 UP000249989 UP000183832 UP000091820 UP000037069 UP000095301 UP000092445 UP000092444
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
Q3C2E3
A0A2A4K4K7
A0A212FF41
A0A023JNR8
A0A1B0YTK4
A0A0N1PHN0
+ More
A0A2J7QC17 A0A2Z5U1Y0 A0A310SHK9 E2B3L1 A0A154P726 F8QQF2 F4X361 E2AIS5 E0VR05 A0A158NPL5 A0A1B6CXK1 A0A0C9QVW1 A0A195FFU9 A0A067RWU2 A0A0J7NCT8 A0A087ZYD6 A0A023EQK1 A0A1Q3FXJ6 Q1L0R1 A0A2J7QC09 U5EY70 A0A151IDR9 A0A1B6FMW4 A0A2H1V829 A0A182JIC7 A0A026WTT7 A0A084WM35 A0A195BEV0 A0A151IVK5 A0A151WEG4 A0A182X017 A0A182US54 A0A182KT27 A0A182HPR1 A0A336M6U7 A0A2H8TSD8 A0A2S2QW05 A0A1B6KES4 A0A182QL11 J9KAX7 A0A2A3E4W9 A0A2M3ZFN3 A0A2M4A467 A0A182FVA0 A0A2M4BS21 G3F9Y2 A0A2M4CTJ6 A0A0K8TPY2 A0A3L8DZU2 A0A1B6EAR8 T1E3S8 A0A165A8C5 A0A0P6IIL7 A0A0M8ZU21 A0A069DTG8 A0A023FAE4 A0A0P6BVV9 E9FQM3 A0A0P5VMX5 A0A1L8DKN4 A0A0P5VMX8 A0A0P4VP85 R4G4C2 A0A1W4WYL0 A0A0V0GAP6 A0A182YC55 A0A1Y1NHD4 Q16YY2 A0A182GRN6 A0A1J1IIP5 L7X0J9 A0A0P5ZL25 A0A0A7D769 A0A2S2NND6 A0A0P4WBX5 A0A023JBY8 A0A0P5SFK8 A0A1B6EUA3 A0A0P5HV78 A0A0P5SSW0 A0A146LVE3 A0A1A9W850 A0A221I025 A0A0P5SPW4 A0A0L0C6N3 A0A0P5DZD2 A0A288ZBA2 A7BHS5 A0A1I8NA24 A0A1L8EC74 A0A1B0A5N9 A0A1B0GB82 A0A2P2I1S0 A0A1B6MB35
A0A2J7QC17 A0A2Z5U1Y0 A0A310SHK9 E2B3L1 A0A154P726 F8QQF2 F4X361 E2AIS5 E0VR05 A0A158NPL5 A0A1B6CXK1 A0A0C9QVW1 A0A195FFU9 A0A067RWU2 A0A0J7NCT8 A0A087ZYD6 A0A023EQK1 A0A1Q3FXJ6 Q1L0R1 A0A2J7QC09 U5EY70 A0A151IDR9 A0A1B6FMW4 A0A2H1V829 A0A182JIC7 A0A026WTT7 A0A084WM35 A0A195BEV0 A0A151IVK5 A0A151WEG4 A0A182X017 A0A182US54 A0A182KT27 A0A182HPR1 A0A336M6U7 A0A2H8TSD8 A0A2S2QW05 A0A1B6KES4 A0A182QL11 J9KAX7 A0A2A3E4W9 A0A2M3ZFN3 A0A2M4A467 A0A182FVA0 A0A2M4BS21 G3F9Y2 A0A2M4CTJ6 A0A0K8TPY2 A0A3L8DZU2 A0A1B6EAR8 T1E3S8 A0A165A8C5 A0A0P6IIL7 A0A0M8ZU21 A0A069DTG8 A0A023FAE4 A0A0P6BVV9 E9FQM3 A0A0P5VMX5 A0A1L8DKN4 A0A0P5VMX8 A0A0P4VP85 R4G4C2 A0A1W4WYL0 A0A0V0GAP6 A0A182YC55 A0A1Y1NHD4 Q16YY2 A0A182GRN6 A0A1J1IIP5 L7X0J9 A0A0P5ZL25 A0A0A7D769 A0A2S2NND6 A0A0P4WBX5 A0A023JBY8 A0A0P5SFK8 A0A1B6EUA3 A0A0P5HV78 A0A0P5SSW0 A0A146LVE3 A0A1A9W850 A0A221I025 A0A0P5SPW4 A0A0L0C6N3 A0A0P5DZD2 A0A288ZBA2 A7BHS5 A0A1I8NA24 A0A1L8EC74 A0A1B0A5N9 A0A1B0GB82 A0A2P2I1S0 A0A1B6MB35
PDB
4ZTH
E-value=1.09345e-152,
Score=1384
Ontologies
PATHWAY
GO
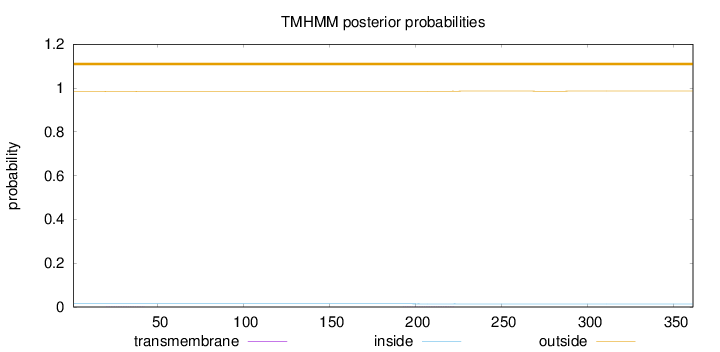
Topology
Length:
361
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0306
Exp number, first 60 AAs:
0.00513
Total prob of N-in:
0.01499
outside
1 - 361
Population Genetic Test Statistics
Pi
205.737242
Theta
168.359783
Tajima's D
0.399181
CLR
0.340902
CSRT
0.48377581120944
Interpretation
Uncertain
